Dr Yeadon’s (former Pfizer VP) Coronavirus Vaccine Safety Petition
Coronavirus vaccine safety concerns
On December 1, 2020, Dr. Michael Yeadon (former Vice President Respiratory & Chief Scientific Advisor, Pfizer) and Dr. Wolfgang Wodarg (lung specialist and former head of the public health department) filed an application with the EMA, the European Medicine Agency responsible for EU-wide drug approval, for the immediate suspension of all SARS CoV 2 vaccine studies, in particular the BioNtech/Pfizer study on BNT162b (EudraCT number 2020-002641-42).
Dr. Wodarg and Dr. Yeadon demand that the studies – for the protection of the life and health of the volunteers – should not be continued until a study design is available that is suitable to address the significant safety concerns expressed by an increasing number of renowned scientists against the vaccine and the study design.
On the one hand, the petitioners demand that, due to the known lack of accuracy of the PCR test in a serious study, a so-called Sanger sequencing must be used. This is the only way to make reliable statements on the effectiveness of a vaccine against Covid-19. On the basis of the many different PCR tests of highly varying quality, neither the risk of disease nor a possible vaccine benefit can be determined with the necessary certainty, which is why testing the vaccine on humans is unethical per se.
Furthermore, they demand that it must be excluded, e.g. by means of animal experiments, that risks already known from previous studies, which partly originate from the nature of the corona viruses, can be realized.
The concerns are directed in particular to the following points:
- The formation of so-called “non-neutralizing antibodies” can lead to an exaggerated immune reaction, especially when the test person is confronted with the real, “wild” virus after vaccination. This so-called antibody-dependent amplification, ADE, has long been known from experiments with corona vaccines in cats, for example. In the course of these studies all cats that initially tolerated the vaccination well died after catching the wild virus.
- The vaccinations are expected to produce antibodies against spike proteins of SARS-CoV-2. However, spike proteins also contain syncytin-homologous proteins, which are essential for the formation of the placenta in mammals such as humans. It must be absolutely ruled out that a vaccine against SARS-CoV-2 could trigger an immune reaction against syncytin-1, as otherwise infertility of indefinite duration could result in vaccinated women.
- The mRNA vaccines from BioNTech/Pfizer contain polyethylene glycol (PEG). 70% of people develop antibodies against this substance – this means that many people can develop allergic, potentially fatal reactions to the vaccination.
- The much too short duration of the study does not allow a realistic estimation of the late effects. As in the narcolepsy cases after the swine flu vaccination, millions of healthy people would be exposed to an unacceptable risk if an emergency approval were to be granted and the possibility of observing the late effects of the vaccination were to follow. Nevertheless, BioNTech/Pfizer apparently submitted an application for emergency approval on December 1, 2020.
December 1, 2020“There is no indication whether antibodies against spike proteins of SARS viruses would also act like anti-Syncytin-1 antibodies.
However, if this were to be the case this would then also prevent the formation of a placenta which would result in vaccinated women essentially becoming infertile.”
Co-sign their petition
Dr. Wodarg and Dr. Yeadon ask as many EU citizens as possible to co-sign their petition by sending the following e-mail prepared by clicking here to the EMA.
Updates
- February 5th, 2021 Open Letter from the UK Medical Freedom Alliance to Nadhim Zahawi (Minister for Covid-19 Vaccine Deployment) and Matt Hancock (Secretary of State for Health and Social Care)
More Resources:
- James Lyons-Weiler — Pathogenic Priming: Coronavirus Vaccine Safety Warning
- Dr. Larry Palevsky — Covid-19 Vaccine Safety Concerns
- Peter Doshi — Is the Coronavirus Vaccine Safe? We Don’t Know Yet
The full petition is below. Those with an even deep interest, the official UK Pfizer/BioNTech vaccine notes may be found here.
PETITIONER: December 1, 2020
Dr. med. Wolfgang Wodarg
Germany
CO-PETITIONER:
Dr. Michael Yeadon
TO:
European Medicines Agency
Committee for human medicinal products (CHMP)
COVID-19 EMA pandemic Task Force (COVID-ETF) Domenico Scarlattilaan 6
1083 HS Amsterdam The Netherlands
!! URGENT !!
PETITION/MOTION FOR ADMINISTRATIVE/REGULATORY ACTION REGARDING
CONFIRMATION OF EFFICACY END POINTS AND USE OF DATA IN CONNECTION WITH THE FOLLOWING CLINICAL TRIAL(S):
PHASE III – EUDRACT NUMBER: 2020-002641-42 SPONSOR PROTOCOL NUMBER: C4591001 SPONSOR:
BIONTECH SE (SOCIETAS EUROPAEA), AN DER GOLDGRUBE 12, 55131 MAINZ, GERMANY
AND ANY OTHER ONGOING CLINICAL TRIALS OF VACCINE CANDIDATES DESIGNED TO STOP TRANSMISSION OF THE VIRUS FROM THE VACCINE RECIPIENT TO OTHERS AND/OR TO PREVENT COVID-19 OR MITIGATE SYMPTOMS OF COVID-19 FOR WHICH PCR RESULTS ARE THE PRIMARY EVIDENCE OF INFECTION
WITH SARS-COV-2 ADMINISTRATIVE/REGULATORY STAY OF ACTION
This petition for a stay of action is submitted by the undersigned (“Petitioner” or “Lead Petitioner”) to request the EMA a) stay the Phase III clinical trial(s) of BNT162b (EudraCT Number 2020-002641-42) in the EU (current protocol country: Germany) until study design is amended to conform with the requests in the “Action Requested” section (B.) below; and b) stay all other clinical trials of vaccine candidates designed to stop transmission of the virus from the
vaccine recipient to others and/or prevent or mitigate symptoms of COVID-19 for which PCR results are the primary evidence of infection.
Because of the compelling need to ensure the safety and efficacy of any COVID-19 vaccine licensed by the EMA (and/or the German Paul-Ehrlich-Institut), and to allow Petitioner the opportunity to seek appropriate emergency judicial relief should the EMA deny its Petition, Petitioner respectfully requests that EMA act on the instant Petition immediately.
A. DECISIONS INVOLVED
I. Approval of trial design and/or decision to not challenge trial design for Phase III trial of BNT162 (EudraCT Number 2020-002641-42)
II. Approval of trial design and/or decision to not challenge trial design of all other clinical trials of vaccine candidates designed to stop transmission of the virus from the vaccine recipient to others and/or to prevent or mitigate symptoms of COVID-19 for which PCR results are the primary evidence of infection.
B. ACTION REQUESTED
I. Stay the Phase III trial of BNT162 in the protocol country Germany and in any other EU protocol countries (as applicable) until study design is amended to provide that:
Before an Emergency Authorization/Conditional Approval and/or Unrestricted Authorization is issued for the Pfizer/BioNTech vaccine, all “endpoints” or COVID-19 cases used to determine vaccine efficacy in the Phase 3 or 2/3 trials should have their infection status confirmed by appropriate Sanger sequencing (as described in section C. III. below), given a) the high cycle thresholds used in some trials; and b) design flaws of certain RT-qPCR tests identical to or modeled after what is sometimes called the “Drosten-Test”.
II. Stay the clinical trials of all vaccine candidates designed to stop transmission of the virus from the vaccine recipient to others and/or to prevent or mitigate symptoms of COVID-19 for which PCR results are the primary evidence of infection until study design is amended to provide that:
Before an Emergency Authorization/Conditional Approval and/or Unrestricted Authorization is issued for vaccine designed to stop transmission of the virus from the vaccine recipient to others and/or to prevent or mitigate symptoms of COVID-19, all “endpoints” or COVID-19 cases used to determine vaccine efficacy should have their infection status confirmed by appropriate Sanger sequencing (as described in section B. III. below), given a) the high cycle thresholds used in some trials; and b) design flaws of certain RT-qPCR tests identical to or modeled after what is sometimes called the “Drosten-Test”.
III. High cycle thresholds, or Ct values, in RT-qPCR test results have been widely acknowledged to lead to false positives. Also, a group of scientists and researchers have recently called for a retraction of the paper that describes the so called “Drosten-Test” (sometimes also being referred to as the “Corman-Drosten protocol” – a specific RT-qPCR assay described by Corman,Victor M., Drosten, Christian and others in “Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR.” Euro Surveillance 2020;25(3):pii=2000045. https://doi.org/10.2807/1560-7917.ES.2020.25.3.2000045).
All RT-qPCR-positive test results used to categorize patient as “COVID-19 cases” in the trials and used to qualify the trial’s endpoints should be verified by Sanger sequencing to confirm that the tested samples in fact contain a unique SARS-CoV-2 genomic RNA. Congruent with FDA and EMA requirements for a confirmed diagnosis of human papillomavirus (HPV) using PCR, the sequencing electropherogram must show a minimum of 100 contiguous bases matching the reference sequence with an Expected Value (E Value) <10-30 for the specific SARS-CoV-2 gene sequence based on a BLAST search of the GenBank database (aka NCBI Nucleotide database).
C. STATEMENT OF GROUNDS
I. As detailed herein, (i) without the requested stay, the Petitioner and many EU residents/citizens will suffer irreparable harm, (ii) the request is not frivolous and is being pursued in good faith, (iii) the request demonstrates sound public policy, and (iv) the public interest favors granting a stay.
II. Petitioner deems the current study designs for the Phase II/III trials of BNT162b (“the Pfizer/BioNTech trial”) to be inadequate to accurately assess efficacy. Petitioner also deems the designs of clinical trials of vaccine candidates designed to stop transmission of the virus from the vaccine recipient to others and/or to prevent or mitigate symptoms of COVID-19 for which PCR results are the primary evidence of infection to be inadequate to accurately assess efficacy.
III. Petitioner and the public will suffer irreparable harm if the actions requested herein are not granted, because once the EMA (and other appropriate bodies in the various EU member states) approves the COVID-19 vaccines in question, both governments of EU member states and employers in the EU are most likely going to recommend them for widespread use. If the assignment of cases and non-cases during the course of the trials is not accurate, the vaccines will not have been properly tested. If the vaccines are not properly tested, important public policy decisions regarding its use will be based on misleading evidence. The medical and economic consequences to EU member states and their residents/citizens could hardly be higher.
IV. Furthermore, if the vaccines are approved without an appropriate and accurate review of efficacy, then any potential acceptance or mandate of these vaccines is likely to be based on inaccurate evidence regarding the vaccine, namely that it will stop transmission of the virus from the vaccine recipient to others and/or that it will reduce COVID-19 disease and deaths. The Pfizer/BioNTech trial protocol and other trial protocols are currently not designed to determine whether either of those objectives can be met; and even if it was, if cases cannot be reliably identified, neither objective could be reliably met.
V. The public interest also weighs strongly in favor of the requested relief because improving the accurate determination of primary endpoints (i) will comport with the best scientific practices, (ii) increase public confidence in the efficacy of a product likely to be mandated or intended for widespread use, and (iii) not doing so will have the opposite result and create uncertainties regarding the efficacy of and need for the COVID-19 vaccines.
VI. Petitioner hereby incorporates the grounds, facts, arguments and opinions stated in the “PETITION FOR ADMINISTRATIVE ACTION REGARDING CONFIRMATION OF EFFICACY END POINTS OF THE PHASE III CLINICAL TRIALS OF COVID-19 VACCINES” which has been submitted to the FDA by Dr. Sin Hang Lee via electronic filing on November 25, 2020 (Exhibit A – Docket No. FDA-2020-P-2225). Exhibit A attached hereto shall be incorporated herein and shall be understood to be a part hereof as though included in the body of this petition.
VII.Petitioner hereby also incorporates the grounds, facts, arguments and opinions stated in the external peer review of the “Drosten- Test” (Exhibit B). Design flaws of certain RT-qPCR tests that are identical to or modeled after what is sometimes called the “Drosten-Test” can lead to false-positive results in trials designed such that PCR results are the primary evidence of infection. Exhibit B attached hereto shall be incorporated herein and shall be understood to be a part hereof as though included in the body of this petition.
VIII. For a vaccine to work, our immune system needs to be stimulated to produce a neutralizing antibody, as opposed to a non-neutralizing antibody. A neutralizing antibody is one that can recognize and bind to some region (‘epitope’) of the virus, and that subsequently results in the virus either not entering or replicating in your cells. A non-neutralizing antibody is one that can bind to the virus, but for some reason, the antibody fails to neutralize the infectivity of the virus. In some viruses, if a person harbors a non-neutralizing antibody to the virus, a subsequent infection by the virus can cause that person to elicit a more severe reaction to the virus due to the presence of the non-neutralizing antibody. This is not true for all viruses, only particular ones. This is called Antibody Dependent Enhancement (ADE), and is a common problem with Dengue Virus, Ebola Virus, HIV, RSV, and the family of coronaviruses. In fact, this problem of ADE is a major reason why many previous vaccine trials for other coronaviruses failed. Major safety concerns were observed in animal models. If ADE occurs in an individual, their response to the virus can be worse than their response if they had never developed an antibody in the first place. This can cause a hyperinflammatory response, a cytokine storm, and a generally dysregulation of the immune system that allows the virus to cause more damage to our lungs and other organs of our body. In addition, new cell types throughout our body are now susceptible to viral infection due to the additional viral entry pathway. There are many studies that demonstrate that ADE is a persistent problem with coronaviruses in general, and in particular, with SARS-related viruses. ADE has proven to be a serious challenge with coronavirus vaccines, and this is the primary reason many of such vaccines have failed in early in-vitro or animal trials. For example, rhesus macaques who were vaccinated with the Spike protein of the SARS-CoV virus demonstrated severe acute lung injury when challenged with SARS-CoV, while monkeys who were not vaccinated did not. Similarly, mice who were immunized with one of four different SARS-CoV vaccines showed histopathological changes in the lungs with eosinophil infiltration after being challenged with SARS-CoV virus.
IX. There are some concerning issues with the trial designs, spelled out by Dr. Peter Doshi in the British Medical Journal. Dr. Doshi focuses on the two biggest issues. First, none of the leading vaccine candidate trials is designed to test if the vaccine can reduce severe COVID-19 symptoms, defined as: hospital admissions, ICU or death. And, second, the trials are not designed to test if the vaccine can interrupt transmission (https://www.bmj.com/content/bmj/371/bmj.m4037.full.pdf). If neither of these conditions is met, the vaccine in essence performs like a therapeutic drug, except a vaccine would be taken prophylactically, even by the perfectly healthy, and more than likely carries a higher risk of injury than a therapeutic drug. If this were to be true, then therapeutic drugs would be superior to any COVID vaccine.
X. In the Pfizer/BioNTech mRNA vaccine candidate, polyethylene glycol (PEG) is found in the fatty lipid nanoparticle coating around the mRNA. Seventy percent of people make antibodies to PEG and most do not know it, creating a concerning situation where many could have allergic, potentially deadly, reactions to a PEG-containing vaccine. PEG antibodies may also reduce vaccine effectiveness. Pfizer/BioNTech is also inserting an ingredient derived from a marine invertebrate, mNeonGreen, into its vaccine. The ingredient has bioluminescent qualities, making it attractive for medical imaging purposes, but it is unclear why an injected vaccine would need to have that quality. mNeonGreen has unknown antigenicity.
XI. Several vaccine candidates are expected to induce the formation of humoral antibodies against spike proteins of SARS-CoV-2. Syncytin-1 (see Gallaher, B., “Response to nCoV2019 Against Backdrop of Endogenous Retroviruses” – http://virological.org/t/response-to-ncov2019- against-backdrop-of-endogenous-retroviruses/396), which is derived from human endogenous retroviruses (HERV) and is responsible for the development of a placenta in mammals and humans and is therefore an essential prerequisite for a successful pregnancy, is also found in homologous form in the spike proteins of SARS viruses. There is no indication whether antibodies against spike proteins of SARS viruses would also act like anti-Syncytin-1 antibodies. However, if this were to be the case this would then also prevent the formation of a placenta which would result in vaccinated women essentially becoming infertile. To my knowledge, Pfizer/BioNTech has yet to release any samples of written materials provided to patients, so it is unclear what, if any, information regarding (potential) fertility-specific risks caused by antibodies isincluded.
According to section 10.4.2 of the Pfizer/BioNTech trial protocol, a woman of childbearing potential (WOCBP) is eligible to participate if she is not pregnant or breastfeeding, and is using an acceptable contraceptive method as described in the trial protocol during the intervention period (for a minimum of 28 days after the last dose of study intervention).
This means that it could take a relatively long time before a noticeable number of cases of post-vaccination infertility could be observed.
XII. It appears that Pfizer/BioNTech have not yet released any samples of written materials provided to patients, so it is unclear what, if any, instructions/information patients/subjects were given regarding ADE and PEG-related issues and (potential) fertility- or pregnancy-specific issues.
D. STAY URGENTLY REQUIRED
I. Petitioner any many EU residents/citizens will suffer irreparable harm because once the EMA approves the COVID- 19 vaccine(s) in question, both governments of EU member states and employers in the EU are most likely going to recommend them for widespread use, and hence without the EMA assuring proper safety trials of the vaccines now, the Petitioner and others will not have the opportunity to object to receiving the vaccine based on deficient clinical trials later.
II. Furthermore, if the vaccines are licensed without an appropriate efficacy review and without improving the accurate determination of primary endpoints, then any potential acceptance or mandate of these vaccines are likely to be based on inaccurate beliefs and data about the vaccines, namely that they will or might stop transmission of the virus from the vaccine recipient to others and/or that it will reduce severe COVID-19 disease and deaths. The trial protocols in question are not currently properly designed to determine whether either of those objectives can be met.
III. This petition is also not frivolous and is being pursued in good faith as it seeks to increase the scientific integrity and reliability of the trials of the COVID-19 vaccines.
IV. Finally, the public interest also weighs strongly in favor of the requested relief because improving the accurate determination of primary endpoints (i) will comport with the best scientific practices, (ii) increase public confidence in the efficacy of a vaccine expected to be mandated or strongly recommended for widespread use, and (iii) not doing so will have the opposite result in that it will create uncertainties regarding the efficacy of and need for the COVID-19 vaccines.
V. The Petitioner therefore respectfully urges that this request be granted
Respectfully submitted on my behalf and on behalf of Co-Petitioner Dr. Michael Yeadon:
![]()
Dr. med. Wolfgang Wodarg
Exhibit A
Exhibit B
Exhibit A
EXPAND
VIA ELECTRONIC FILING
November 25, 2020
Division of Dockets Management Department of Health and Human Services Food and Drug Administration Commissioner Stephen M. Hahn, M.D.
5630 Fishers Lane
Rm. 1061
Rockville, MD 20852
UNITED STATES DEPARTMENT OF HEALTH AND HUMAN SERVICES AND THE FOOD AND DRUG ADMINISTRATION
PETITION FOR ADMINISTRATIVE :
ACTION REGARDING :
CONFIRMATION OF EFFICACY :
END POINTS OF THE PHASE III : Docket No. FDA-2020-P-2225 CLINICAL TRIALS OF COVID-19 :
VACCINES :
ADMINISTRATIVE STAY OF ACTION
This petition for a stay of action is submitted on behalf of Dr. Sin Hang Lee (“Petitioner”) pursuant to 21 C.F.R. § 10.35 and related and relevant provisions of the Federal Food, Drug, and Cosmetic Act or the Public Health Service Act to request the Commissioner of Food and Drugs (the “Commissioner”) stay the Phase III trials of BNT162b (NCT04368728) to conform with the requests in the “Action Requested” section below.
Because of the compelling need to ensure the safety and efficacy of any COVID-19 vaccine licensed by the FDA, and to allow Petitioner the opportunity to seek emergency judicial relief should the Commissioner deny its Petition, Petitioner respectfully requests that FDA act on the instant Petition by December 11, 2020.
A. DECISION INVOLVED
- Approval of trial design for Phase III trial of BNT162 (NCT04368728)1
1 NCT04368728 available at https://www.clinicaltrials.gov/ct2/show/NCT04368728 (last visited November 3, 2020).
B. ACTION REQUESTED
- Stay the Phase III trial of BNT162 (NCT04368728) until its study design is amended to provide that:
Before an EUA or unrestricted license is issued for the Pfizer vaccine, or for other vaccines for which PCR results are the primary evidence of infection, all “endpoints” or COVID-19 cases used to determine vaccine efficacy in the Phase 3 or 2/3 trials should have their infection status confirmed by Sanger sequencing, given the high cycle thresholds used in some trials. High cycle thresholds, or Ct values, in RT-qPCR test results have been widely acknowledged to lead to false positives.2
All RT-qPCR-positive test results used to categorize patient as “COVID-19 cases” and used to qualify the trial’s endpoints should be verified by Sanger sequencing to confirm that the tested samples in fact contain a unique SARS-CoV-2 genomic RNA. Congruent with FDA requirements for a confirmed diagnosis of human papillomavirus (HPV) using PCR, the sequencing electropherogram must show a minimum of 100 contiguous bases matching the reference sequence with an Expected Value (E Value) <10-30 for the specific SARS-CoV-2 gene sequence based on a BLAST search of the GenBank database (aka NCBI Nucleotide database).
C. STATEMENT OF GROUNDS
- As detailed herein, (i) without the requested stay, the Petitioner will suffer irreparable harm, (ii) the request is not frivolous and is being pursued in good faith, (iii) the request demonstrates sound public policy, and (iv) the public interest favors granting a stay. 3
- The current study designs for the Phase II/III trials of BNT162b (“the Pfizer Vaccine”) are inadequate to accurately assess efficacy.
- Petitioner and the public will suffer irreparable harm if the actions requested herein are not granted, because once the FDA licenses this COVID-19 vaccine, both governments and employers may make this product mandatory (in general, or for airline or international travel) or may recommend it for widespread use. If the assignment of cases and non-cases during the course of the trial is not accurate, the vaccine will not have been properly If the vaccine is not properly tested, important public policy decisions regarding its use will be based on misleading evidence. The medical and economic consequences to the nation could hardly be higher.
2 See New York Times. Your Coronavirus Test Is Positive. Maybe It Shouldn’t Be. By Apoorva Mandavilli. Published Aug. 29, 2020 and updated Sept. 17, 2020, available at https://www.nytimes.com/2020/08/29/health/coronavirus- testing.html.
3 The Petitioner hereby incorporates by reference as if fully set forth herein the Statement of Grounds from its Citizen’s Petition, dated November 23, 2020, available at, https://beta.regulations.gov/document/FDA-2020-P-2225 (last visited November 25, 2020).
- The New York State Bar Association has already issued a report on COVID-19 recommending that, “a vaccine subject to scientific evidence of safety and efficacy be made widely available, and widely encouraged, and if the public health authorities conclude necessary, required…”4 Thus, it is reasonable to suspect that COVID-19 vaccines, including the Pfizer vaccine, could become mandatory. Without the FDA assuring proper efficacy trials of the vaccine now, the Petitioner and the public may not have the opportunity to object to receiving the vaccine, which was approved based on currently deficient and unreliable clinical trial data.
- Furthermore, if the vaccine is approved without an appropriate and accurate review of efficacy, then any potential acceptance or mandate of these vaccines is likely to be based on inaccurate evidence regarding the vaccine, namely that it will stop transmission of the virus from the vaccine recipient to others and/or that it will reduce severe COVID-19 disease and deaths. The Pfizer trial protocol is currently not designed to determine whether either of those objectives can be met; and even if it was, if cases cannot be reliably identified, neither objective could be reliably met.
- The public interest also weighs strongly in favor of the requested relief because improving the accurate determination of primary endpoints (i) will comport with the best scientific practices, (ii) increase public confidence in the efficacy of a product likely to be mandated or intended for widespread use, and (iii) not doing so will have the opposite result and create uncertainties regarding the efficacy of and need for the COVID-19 vaccines.
- According to the trial protocol, “8.1. Efficacy and/or Immunogenicity Assessments,” the trial’s primary endpoint is prevention of symptomatic disease in vaccine recipients. In order to evaluate that endpoint, the trial will track recorded COVID-19 disease. The definition of confirmed COVID-19 is:
presence of at least 1 of the following symptoms and SARS-CoV-2 NAAT-positive during, or within 4 days before or after, the symptomatic period, either at the central laboratory or at a local testing facility (using an acceptable test):
- Fever;
- New or increased cough;
- New or increased shortness of breath;
- Chills;
- New or increased muscle pain;
- New loss of taste or smell;
- Sore throat;
- Diarrhea;
- Vomiting
4 https://nysba.org/app/uploads/2020/06/2b-REV-6-12-20-FINAL-HOD-RESOLUTIONS-1-through-4.pdf.
- As a result, if a participant has a positive reverse transcription-quantitative polymerase chain reaction (“RT-qPCR”) test along with a cough or sore throat, that participant would be considered as a “confirmed COVID-19 case” and would be counted as an endpoint. Once a trial reaches a certain number of “endpoints”, the trial is closer to seeking FDA approval or licensure by demonstrating that the vaccine is “effective” (in that the vaccine group had lower incidence of endpoints than the control group).
- This effectively means that the efficacy of the vaccine will be determined based on only symptoms of non-specific disease in conjunction with a PCR positive laboratory test.
- According to the trial protocol, “8.1 Efficacy and/or Immunogenicity Assessments,” efficacy will be assessed throughout a participant’s involvement in the study through surveillance for potential cases of COVID-19. If, at any time, a participant develops acute respiratory illness (see Section 8.13), for the purposes of the study he or she will be considered to potentially have COVID-19 illness. In this circumstance, the participant should contact the site, an in-person or telehealth visit should occur, and assessments should be conducted as specified in the SoA. The assessments will include a nasal (midturbinate) swab, which will be tested at a central laboratory using a reverse transcription–polymerase chain reaction (RT-PCR) test (Cepheid; FDA approved under EUA), or other equivalent nucleic acid amplification–based test (ie, NAAT), to detect SARS-CoV-2. In addition, clinical information and results from local standard-of-care tests (as detailed in Section 8.13) will be assessed. The central laboratory NAAT result will be used for the case definition, unless no result is available from the central laboratory, in which case a local NAAT result may be used if it was obtained using 1 of the following assays:
- Cepheid Xpert Xpress SARS-CoV-2
- Roche cobas SARS-CoV-2 real-time RT-PCR test (EUA200009/A001)
- Abbott Molecular/RealTime SARS-CoV-2 assay (EUA200023/A001)
- These test kits referred to in the trial protocol, namely the Cepheid Xpert Xpress SARS-CoV-2, the Roche cobas SARS-CoV-2 real-time RT-PCR test (EUA200009/A001), and the Abbott Molecular/RealTime SARS-CoV-2 assay (EUA200023/A001), are very unreliable tools when they are used to determine whether the nasal swab sample collected from a symptomatic participant contains SARS-CoV-2 or not. These real-time RT-PCR or RT- quantitative PCR tests should be referred to as rRT-PCR or RT-qPCR tests to be distinguished from conventional RT-PCR. The very short RT-qPCR product (amplicon) cannot be analyzed by automated Sanger sequencing as the products of conventional PCR can. And DNA sequencing for validation of the PCR products is needed to correctly determine if the presumptive RT-qPCR- positive SARS-CoV-2 test result is a true positive or a false positive. The reasoning is further outlined as follows:
a. Nowadays DNA sequencing of the PCR amplicon of the genomic nucleic acid of the pathogen is a universally accepted technology for detection and for confirmation of infectious agents, especially pathogenic viruses, in clinical specimens. On January 10,2020, the first SARS-CoV-2 genome sequence was released online. On the same day, a group of American scientists, most from the CDC, immediately designed 2 complementary panels of primers to amplify the virus genome for sequencing. The PCR amplicons averaged 550 bp in size in their research.5
b. The World Health Organization (WHO) guidance titled “WHO Laboratory testing for coronavirus disease (COVID-19) in suspected human cases-Interim guidance dated 19 March 2020” advised “Routine confirmation of cases of COVID-19 is based on detection of unique sequences of virus RNA by NAAT such as real-time reverse transcription- polymerase chain reaction (rRT-PCR) with confirmation by nucleic acid sequencing when necessary.”6
c. The FDA also recognizes the inherent inaccuracy of the RT-qPCR tests. In its letter issued on February 4, 2020 authorizing emergency use of the CDC 2019-Novel Coronavirus (2019-nCoV, renamed as SARS-CoV-2) Real-Time Reverse Transcriptase (RT)-PCR Diagnostic Panel, the FDA specifically stated that the test panel is “for the presumptive qualitative detection of nucleic acid from the 2019-nCoV (sic) in upper and lower respiratory specimens.”7
d. In addition to false-negative results, these RT-qPCR test kits under EUA also generate false-positive test results. For example, 77 positive SARS-CoV-2 test results on a group of football players all turned out to be false positives on repeat test.8
e. The FDA has officially alerted clinical laboratory staff and health care providers of an increased risk of false-positive results with some of these commercial test kits permitted to be used under EUA.9
5 Paden CR, Tao Y, Queen K, Zhang J, Li Y, Uehara A, Tong S. Rapid, Sensitive, Full-Genome Sequencing of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg Infect Dis. 2020 Oct;26(10):2401-2405. doi: 10.3201/eid2610.201800. Epub 2020 Jul 1. PMID: 32610037; PMCID: PMC7510745.
6 WHO Laboratory testing for coronavirus disease (COVID-19) in suspected human cases-Interim guidance 19 March 2020. Available from: https://www.who.int/publications/i/item/10665-331501.
7 FDA letter dated February 4, 2020 authorizing emergency use of the CDC 2019-Novel Coronavirus (2019-nCoV, renamed as SARS-CoV-2) Real-Time Reverse Transcriptase (RT)-PCR Diagnostic Panel. See Open letter from FDA to Robert R. Redfield, MD, Director, Centers for Disease Control and Prevention. March 15, 2020. https://www.fda.gov/media/134919/download.
8 Kevin Patra. Around the NFL- All 77 false-positive COVID-19 tests come back negative upon reruns. Aug 24, 2020. Available from: https://www.nfl.com/news/all-77-false-positive-covid-19-tests-come-back-negative-upon-reruns.
9 FDA. False Positive Results with BD SARS-CoV-2 Reagents for the BD Max System – Letter to Clinical Laboratory Staff and Health Care Providers. Available from: https://www.fda.gov/medical-devices/letters-health-care- providers/false-positive-results-bd-sars-cov-2-reagents-bd-max-system-letter-clinical-laboratory-staff-and Accessed November 2, 2020; see also FDA. Risk of Inaccurate Results with Thermo Fisher Scientific TaqPath COVID-19 Combo Kit – Letter to Clinical Laboratory Staff and Health Care Providers. Available from: https://www.fda.gov/medical-devices/letters-health-care-providers/risk-inaccurate-results-thermo-fisher-scientific- taqpath-covid-19-combo-kit-letter-clinical?utm_campaign=2020-08-17%20Risk%20of%20Inaccurate%20Results
%20with%20Thermo%20Fisher%20Scientific%20TaqPath&utm_medium=email&utm_source=Eloqua.
f. To resolve the problems caused by these inherently inaccurate tests, the FDA’s position is that false results can be investigated using an additional EUA RT-qPCR assay, and/or Sanger sequencing.10 Since an additional EUA RT-qPCR test result may also generate a false result, Sanger sequencing is the de facto gold standard for confirmation of presumptive qualitative detection of nucleic acid from the SARS-CoV-2 and for excluding false-positive cases.
g. According to the FDA guidance on molecular diagnosis of viral infection caused by human papillomavirus (HPV), a conventional PCR detection of genomic DNA followed by Sanger sequencing on both strands of the PCR amplicon (bi-directional sequencing) that contains a minimum of 100 contiguous bases is acceptable as valid diagnostics for HPV infection provided the sequence matches the reference or consensus sequence, e.g. with an Expected Value (E Value) <10-30 for the specific HPV DNA target based on a BLAST search of the GenBank (NCBI Nucleotide) database.11 Following this FDA guidance, and showing the feasibility of implementing the FDA guidance for accurate diagnosis of COVID-19, a protocol using the nested PCR cDNA amplicon of a 398-base highly conserved SARS- CoV-2 N gene segment as the template for Sanger sequencing was developed for confirmatory detection of SARS-CoV-2 in clinical samples. 12
h. DNA sequencing verification is necessary for confirmation of the presumptive SARS- CoV-2-positive cases in the Pfizer vaccine’s Phase II/III clinical trial because, according to its Protocol, the specimens collected from the symptomatic trial subjects were sent to a central laboratory using a reverse transcription–polymerase chain reaction (RT-PCR) test (Cepheid; FDA approved under EUA), or other equivalent nucleic acid amplification– based test (i.e., NAAT), to detect SARS-CoV-2.
In order to raise the detection sensitivity, the mean Ct value of the Cepheid system is set as high as 42.9 for the N2 target, and as high as 44.9 for the E target, as shown in Table 4 of Instructions for Users (Cepheid 302-3562, Rev. E September 2020).13
10 FDA. Molecular Diagnostic Template for Laboratories. Policy for Coronavirus Disease-2019 Tests During the Public Health Emergency (Revised) Available from: https://www.fda.gov/media/135659/download .
11 FDA. Establishing the Performance Characteristics of In Vitro Diagnostic Devices for the Detection or Detection and Differentiation of Human Papillomaviruses. Available from: https://www.fda.gov/media/92930/download.
12 Lee SH. Testing for SARS-CoV-2 in cellular components by routine nested RT-PCR followed by DNA sequencing. International Journal of Geriatrics and Rehabilitation. 2020; 2:69-96. Available from: http://www.int-soc-clin- geriat.com/info/wpcontent/uploads/2020/03/Dr.-Lees-paper-on-testing-for-SARS-CoV-2.pdf.
13 Cepheid. GeneXpert. Instructions for Users. XPRSARS-COV2-10. 302-3562, Rev. E September 2020 https://www.cepheid.com/Package%20Insert%20Files/Xpress-SARS-CoV-2/Xpert%20Xpress%20SARS-CoV- 2%20Assay%20ENGLISH%20Package%20Insert%20302-3562-GX%20Rev.%20E.pdf.
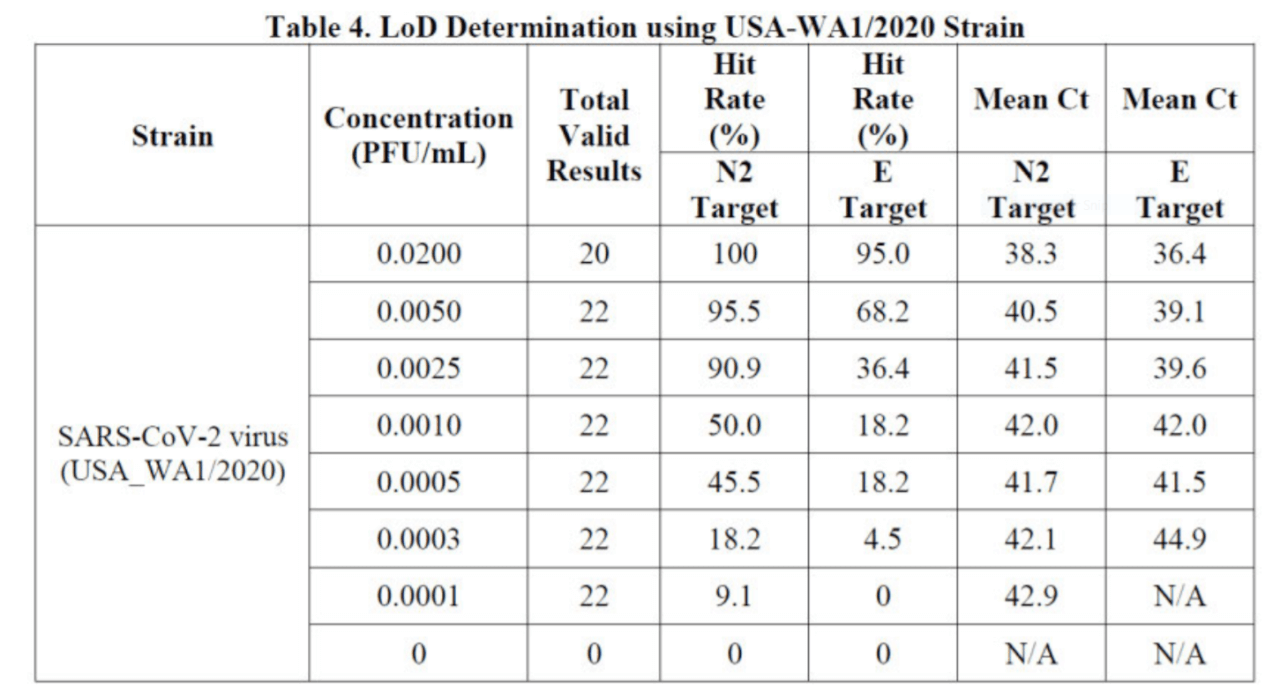
At Ct values between 36.0 and 44.9, many RT-qPCR positive test results are false positives.
i. The results of the 3 RT-qPCR test kits used in the trial protocol are not comparable. A sample identified as negative by the Abbott kit can be classified as positive by the Cepheid kit. According to an FDA survey, the limit of detection by the Cepheid Xpert Xpress SARS-CoV-2 test kit and the limit of detection by Abbott RealTime SARS-CoV-2 assay kit are found to be identical, namely both being at 5400 NAAT Detectable Units/ mL, as shown in the comparative data extracted from an FDA reference panel. 14
| 5400 | Cepheid | Xpert Xpress SARS-CoV-2 test |
| 5400 | Abbott Molecular | Abbott RealTime SARS-CoV-2 assay |
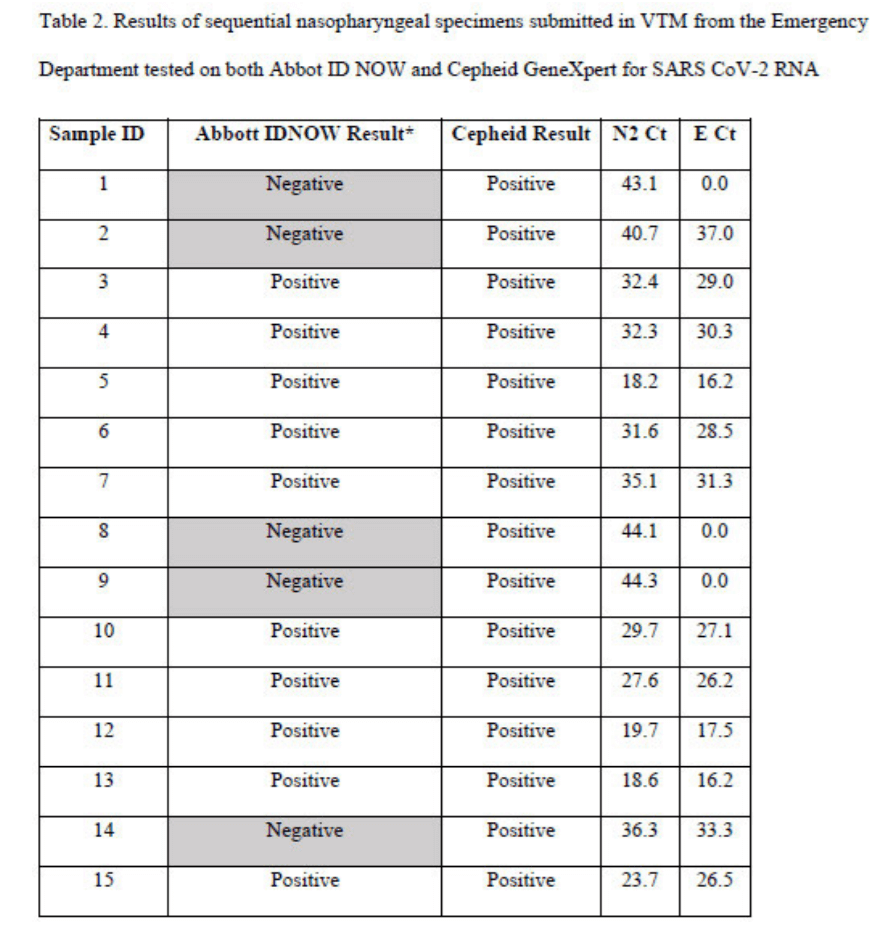
However, due to the designation of higher cycle threshold test results as positives, the Cepheid Xpert kits have classified many Abbott kit negative cases as positives in a head- to-head comparative study as shown in the following “Table 2” extracted from a report by Basu et al.15
14 FDA. SARS-CoV-2 Reference Panel Comparative Data. https://www.fda.gov/medical-devices/coronavirus-covid- 19-and-medical-devices/sars-cov-2-reference-panel-comparative- data.
15 See bioRxiv preprint doi: https://doi.org/10.1101/2020.05.11.089896; Basu A, Zinger T, Inglima K, Woo KM, Atie O, Yurasits L, See B, Aguero-Rosenfeld ME. Performance of Abbott ID Now COVID-19 Rapid Nucleic Acid Amplification Test Using Nasopharyngeal Swabs Transported in Viral Transport Media and Dry Nasal Swabs in a New York City Academic Institution. J Clin Microbiol. 2020 Jul 23;58(8):e01136-20. doi: 10.1128/JCM.01136-20. PMID: 32471894; PMCID: PMC7383552.
j. One of the Cepheid Xpert kit users has put out an alert, stating “The instruments are presently set by the manufacturer to interpret a single target positive with very poor amplification efficiency (high Cycle Threshold [Ct] and/or atypical curve) as ‘DETECTED.’ None of these to date have confirmed positive when tested on other systems using similar targets, and may be a false positive due to background noise.”16
k. Another group of users also found that some tested samples classified as positives by the Cepheid test kits cannot be confirmed with other test kits. These authors published a report, stating: “We found that the sensitivity of the Xpert Xpress SARS-CoV-2 assay was 100% (20 of 20) and the specificity was 80% (16 of 20). When looking at the cycle threshold (Ct) values from the GeneXpert assay we observed that specimens with no amplification of the E gene (ie, Ct=0) and Ct values for the N2 gene greater than 40 cycles were considered as positives, whereas they were negative using the other RT-PCR system (Da An Gene).”17
16 Diagnostic Laboratory Services Inc. Technical Alert. Cepheid GeneXpert and BD Max Instruments may be Reporting False Positives. https://dlslab.com/documents/bulletins/2020/tech-memo-sars-cov-2-pcr-possible-false- positive-6-19-2020.pdf.
17 Rakotosamimanana N, Randrianirina F, Randremanana R, Raherison MS, Rasolofo V, Solofomalala GD, Spiegel A, Heraud JM. GeneXpert for the diagnosis of COVID-19 in LMICs. Lancet Glob Health. 2020 Oct 19:S2214- 109X(20)30428-9. doi: 10.1016/S2214-109X(20)30428-9. Epub ahead of print. PMID: 33091372; PMCID: PMC7572106.
- DNA sequencing verification of the RT-qPCR positive test results is absolutely necessary in this placebo-controlled randomized clinical trial because de facto unblinding has occurred among the participants. According to the trial protocol Section 8.13. COVID-19 Surveillance (All Participants), “If a participant experiences any of the following (irrespective of perceived etiology or clinical significance), he or she is instructed to contact the site immediately and, if confirmed, participate in an in-person or telehealth visit as soon as possible.” This contact would trigger an automatic NAAT test by a Cepheid RT-qPCR assay at the central laboratory or at a local laboratory by any similar acceptable methods.
At the time of enrollment, the participants were informed that each of them would be injected with a vaccine to protect against COVID-19 infection or a saline placebo without disclosing which one of the two was injected into the participant. However, all participants were also informed that the vaccine may cause the following reactions:
- Fever ≤39.0°C (≤102.1°F).
- Redness or swelling at the injection site measuring greater than 10cm (>20 measuring device units).
- Severe pain at the injection site.
- Any severe systemic
It is commonly known to the general public and especially to the informed clinical trial participants that intramuscular injection of a very small amount of sterile normal saline will not cause fever, local redness and swelling, and severe pain, or systemic reactions. The participants receiving placebo would intuitively or reasonably know that they were not injected with a vaccine and were not protected against COVID-19 disease due to the lack of any vaccine reaction after the injection. As a result, more participants receiving placebo than those receiving vaccine would report to the “site” manager when they developed any minor symptoms, such as a sore throat or a new cough for the fear of coming down with COVID-19. The site manager must investigate the symptoms reported, including ordering a RT-qPCR test by Cepheid assay to be performed at the Central Laboratory according to Protocol. The more severe cases might be tested locally by Abbott kits or Roche kits because they might have to be tested in the hospital after admission, and because many hospitals are aware of the high false positive rates generated by the Cepheid kits. The results generated by these test kits are not comparable since the Cepheid test kits using a very high Ct value up to 44.9 for “detection of SARS-CoV-2 genomic RNA” tend to generate many more false positives than the other test kits. A higher number of false-positive test results in the participants receiving placebo will artificially raise the efficacy of the vaccine, unless the RT- qPCR test results are verified by nucleotide sequencing to eliminate all false-positive test results.
- Based on an MPR report published on November 8, 2020, there are only 180 confirmed cases of COVID-19 in this clinical trial series that have been analyzed to support the vaccine efficacy evaluation.18 If the Sponsor (BioNTech/Pfizer) is unable to perform confirmatory Sanger sequencing tests on these 180 RNA extract residual samples, the Petitioner hereby offers to re-test them immediately with Sanger sequencing19 and submit the laboratory data to support FDA’s evaluation. Therefore, there is no excuse for the Sponsor to refuse using the gold standard Sanger sequencing technology for endpoint validation.
18 Diana Ernst, RPh. Final Analysis Reveals COVID-19 Vaccine Candidate BNT162b2 95% Effective. MPR Report. November 18, 2020. https://www.empr.com/home/news/drugs-in-the-pipeline/pfizer-biontech-mrna-based-vaccine- bnt162b2-against-covid19-effective/.
- In summary, based on the scientific data available in the public domain and the FDA guidance, all RT-qPCR test results for detection of SARS-CoV-2 gene sequence must be considered presumptive. The Cepheid test kits for SARS-CoV-2 are known to generate more false- positive test results than other EUA assay kits.
- The residues of the tested samples that were classified as positive for SARS-CoV- 2 by the Cepheid GeneXpert assay, or equivalent as stated in the Pfizer Clinical Trial Protocol, must be re-tested by a Sanger sequencing method to confirm that the presumptive positive samples in fact contain a unique sequence of SARS-CoV-2 genome. Only then can the positive test results from the Cepheid GeneXpert test kits be accepted as an accurate component of the “endpoint.” Only then can one nonspecific symptom plus laboratory positivity be accepted as a valid measure of confirmed COVID-19 cases or “endpoints.”
Stay Urgently Required
- Petitioner will suffer irreparable harm because once the FDA licenses this COVID- 19 vaccine, states are expected to make this product mandatory, and hence without the FDA assuring proper safety trials of the vaccine now, the Petitioner will not have the opportunity to object to receiving the vaccine based on deficient clinical trials later.
- For example, the New York State Bar Association recently passed a resolution recommending that “[s]hould the level of immunity be deemed insufficient by expert medical and scientific consensus to check the spread of COVID-19 and reduce morbidity and mortality, a mandate and state action should be considered.”20 Mandating administration of the vaccine, thereby eliminating the right to informed consent, makes acute the need to assure that the safety and efficacy of any COVID-19 vaccine is robustly studied in an adequate clinical trial monitoring for any potential adverse events.
- Furthermore, if the vaccine is licensed without an appropriate efficacy review and without improving the accurate determination of primary endpoints, then any potential acceptance or mandate of these vaccines are likely to be based on inaccurate beliefs about the vaccine, namely that it will stop transmission of the virus from the vaccine recipient to others or that it will reduce severe COVID-19 disease and deaths. The trial protocols are not currently designed to determine whether either of those objectives can be met.
19 Lee SH. Testing for SARS-CoV-2 in cellular components by routine nested RT-PCR followed by DNA sequencing. International Journal of Geriatrics and Rehabilitation. 2020; 2:69-96. Available from: http://www.int-soc-clin- geriat.com/info/wpcontent/uploads/2020/03/Dr.-Lees-paper-on-testing-for-SARS-CoV-2.pdf.
20 https://nysba.org/app/uploads/2020/11/11.-Health-Law-Section-COVID-19-Report-September-20-2020-with-all- comments.pdf (emphasis added) (last visited November 10, 2020).
- This request is also not frivolous and is being pursued in good faith as it seeks to increase the scientific integrity and reliability of the trials of the COVID-19 Vaccines.
- Finally, the public interest also weighs strongly in favor of the requested relief because improving the accurate determination of primary endpoints (i) will comport with the best scientific practices, (ii) increase public confidence in the efficacy of a product expected to be mandated, and (iii) not doing so will have the opposite result in that it will create uncertainties regarding the efficacy of and need for the COVID-19 Vaccines.
- The Petitioner therefore respectfully urges that this request be granted
Respectfully submitted,
![]()
Dr. Sin Hang Lee
Exhibit B
EXPANDExternal peer review of the RTPCR test to detect SARS-CoV-2 reveals 10 major scientific flaws at the molecular and methodological level: consequences for false positive results.
Pieter Borger(1 * ), Bobby Rajesh Malhotra(2) , Michael Yeadon(3) , Clare Craig(4) Kevin McKernan(5) , Klaus Steger(6) , Paul McSheehy(7) , Lidiya Angelova(8) Fabio Franchi(9), Thomas Binder(10), Henrik Ullrich(11) , Makoto Ohashi(12) Stefano Scoglio(13), Marjolein Doesburg−van Kleffens(14), Dorothea Gilbert(15) Rainer Klement(16), Ruth Schruefer(17), Berber W. Pieksma(18), Jan Bonte(19) Bruno H. Dalle Carbonare(20), Kevin P. Corbett(21), Ulrike Kämmerer(22)
* Corresponding Author
ABSTRACT
“In the publication entitled “Detection of 2019 novel coronavirus (2019−nCoV) by real−time RT−PCR” (Eurosurveillance 25(8) 2020) the authors present a diagnostic workflow and RT−qPCR protocol for detection and diagnostics of 2019−nCoV (now known as SARS−CoV−2), which they claim to be validated, as well as being a robust diagnostic methodology for use in public−health laboratory settings.
In light of all the consequences resulting from this very publication for societies worldwide, a group of independent researchers performed a point−by−point review of the aforesaid publication in which
1) all components of the presented test design were cross checked, 2) the RT−qPCR protocol−recommendations were assessed with respect to good laboratory practice, and 3) parameters examined against relevant scientific literature covering the field.
The published RT−qPCR protocol for detection and diagnostics of 2019−nCoV and the manuscript suffer from numerous technical and scientific errors, including insufficient primer design, a problematic and insufficient RT−qPCR protocol, and the absence of an accurate test validation. Neither the presented test nor the manuscript itself fulfils the requirements for an acceptable scientific publication. Further, serious conflicts of interest of the authors are not mentioned. Finally, the very short timescale between submission and acceptance of the publication (24 hours) signifies that a systematic peer review process was either not performed here, or of problematic poor quality.
We provide compelling evidence of several scientific inadequacies, errors and flaws. Considering the scientific and methodological blemishes presented here, we are confident that the editorial board of Eurosurveillance has no other choice but to retract the publication.”
CONCISE REVIEW REPORT
This paper will show numerous serious flaws in the Corman−Drosten paper, the significance of which has led to worldwide misdiagnosis of infections attributed to SARS−CoV−2 and associated with the disease COVID−19. We are confronted with stringent lockdowns which have destroyed many people’s lives and livelihoods, limited access to education and these imposed restrictions by governments around the world are a direct attack on people’s basic rights and their personal freedoms, resulting in collateral damage for entire economies on a global scale.
There are ten fatal problems with the Corman-Drosten paper which we will outline and explain in greater detail in the following sections.
The first and major issue is that the novel Coronavirus SARS−CoV−2 (in the publication named 2019−nCoV and in February 2020 named SARS−CoV−2 by an international consortium of virus experts) is based on in silico (theoretical) sequences, supplied by a laboratory in China [1], because at the time neither control material of infectious (“live”) or inactivated SARS−CoV−2 nor isolated genomic RNA of the virus was available to the authors. To date no validation has been performed by the authorship based on isolated SARS−CoV−2 viruses or full length RNA thereof. According to Corman et al.:
“We aimed to develop and deploy robust diagnostic methodology for use in public health laboratory settings without having virus material available.” [1]
The focus here should be placed upon the two stated aims: a) development and b) deployment of a diagnostic test for use in public health laboratory settings. These aims are not achievable without having any actual virus material available (e.g. for determining the infectious viral load). In any case, only a protocol with maximal accuracy can be the mandatory and primary goal in any scenario−outcome of this magnitude. Critical viral load determination is mandatory information, and it is in Christian Drosten’s group responsibility to perform these experiments and provide the crucial data.
Nevertheless these in silico sequences were used to develop a RT−PCR test methodology to identify the aforesaid virus. This model was based on the assumption that the novel virus is very similar to SARS−CoV from 2003 as both are beta−coronaviruses.
The PCR test was therefore designed using the genomic sequence of SARS−CoV as a control material for the Sarbeco component; we know this from our personal email−communication with [2] one of the co−authors of the Corman−Drosten paper. This method to model SARS−CoV−2 was described in the Corman−Drosten paper as follows:
“the establishment and validation of a diagnostic workflow for 2019−nCoV screening and specific confirmation, designed in absence of available virus isolates or original patient specimens. Design and validation were enabled by the close genetic relatedness to the 2003 SARS−CoV, and aided by the use of synthetic nucleic acid technology.”
The Reverse Transcription−Polymerase Chain Reaction (RT−PCR) is an important biomolecular technology to rapidly detect rare RNA fragments, which are known in advance. In the first step, RNA molecules present in the sample are reverse transcribed to yield cDNA. The cDNA is then amplified in the polymerase chain reaction using a specific primer pair and a thermostable DNA polymerase enzyme. The technology is highly sensitive and its detection limit is theoretically 1 molecule of cDNA. The specificity of the PCR is highly influenced by biomolecular design errors.
What is important when designing an RT-PCR Test and the quantitative RT-qPCR test described in the Corman-Drosten publication?
1. The primers and probes:
- the concentration of primers and probes must be of optimal range (100−200 nM)
- must be specific to the target−gene you want to amplify
- must have an optimal percentage of GC content relative to the total nitrogenous bases (minimum 40%, maximum 60%)
- for virus diagnostics at least 3 primer pairs must detect 3 viral genes (preferably as far apart as possible in the viral genome)
2. The temperature at which all reactions take place:
- DNA melting temperature (>92˚)
- DNA amplification temperature (TaqPol specific)
- Tm; the annealing temperature (the temperature at which the primers and probes reach the target bindingƒdetachment, not to exceed 2 ̊C per primer pair). Tm heavily depends on GC content of the primers
3. The number of amplification cycles (less than 35; preferably 25-30 cycles);
In case of virus detection, >35 cycles only detects signals which do not correlate with infectious virus as determined by isolation in cell culture [reviewed in 2]; if someone is tested by PCR as positive when a threshold of 35 cycles or higher is used (as is the case in most laboratories in Europe & the US), the probability that said person is actually infected is less than 3%, the probability that said result is a false positive is 97% [reviewed in 3]
- Molecular biological validations; amplified PCR products must be validated either by running the products in a gel with a DNA ruler, or by direct DNA sequencing
- Positive and negative controls should be specified to confirm/refute specific virus detection
- There should be a Standard Operational Procedure (SOP) available
SOP unequivocally specifies the above parameters, so that all laboratories are able to set up the exact same test conditions. To have a validated universal SOP is essential, because it enables the comparison of data within and between countries.
MINOR CONCERNS WITH THE CORMAN-DROSTEN PAPER
- In Table 1 of the Corman−Drosten paper, different abbreviations are stated − “nM” is specified, “nm” isn’t. Further in regards to correct nomenclature, nm means “nanometer” therefore nm should read nM
- It is the general consensus to write genetic sequences always in the 5’−3’ direction, including the reverse primers. It is highly unusual to do alignment with reverse complementary writing of the primer sequence as the authors did in figure 2 of the Corman−Drosten paper. Here, in addition, a wobble base is marked as “y” without description of the bases the Y stands
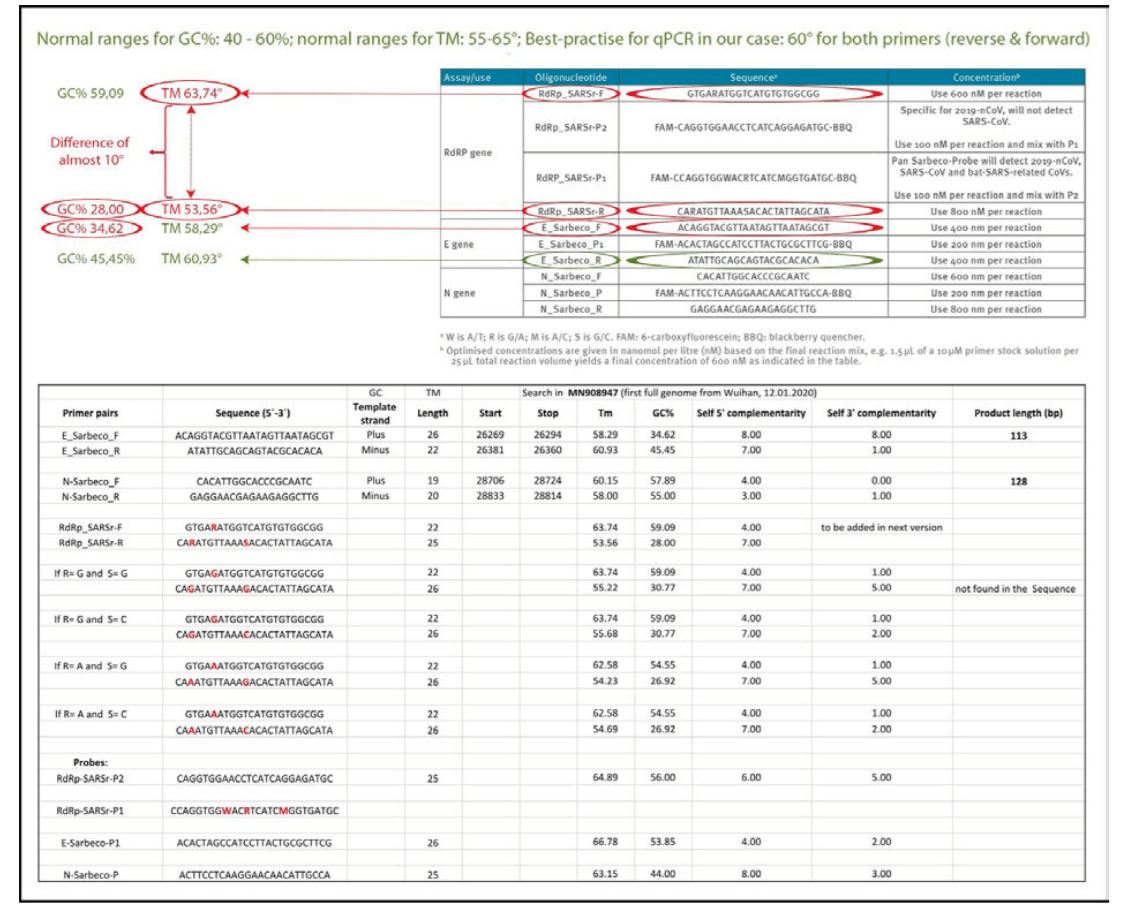
- Two misleading pitfalls in the Corman−Drosten paper are that their Table 1 does not include Tm−values (annealing−temperature values), neither does it show GC−values (number of G and C in the sequences as %−value of total bases).
MAJOR CONCERNS WITH THE CORMAN-DROSTEN PAPER
A) BACKGROUND
The authors introduce the background for their scientific work as: “The ongoing outbreak of the recently emerged novel coronavirus (2019−nCoV) poses a challenge for public health laboratories as virus isolates are unavailable while there is growing evidence that the outbreak is more widespread than initially thought, and international spread through travelers does already occur”.
According to BBC News [4] and Google Statistics [5] there were 6 deaths world−wide on January 21st 2020 − the day when the manuscript was submitted. Why did the authors assume a challenge for public health laboratories while there was no substantial evidence at that time to indicate that the outbreak was more widespread than initially thought?
As an aim the authors declared to develop and deploy robust diagnostic methodology for use in public health laboratory settings without having virus material available. Further, they acknowledge that “The present study demonstrates the enormous response capacity achieved through coordination of academic and public laboratories in national and European research networks.”
B) METHODS AND RESULTS
1. Primer & Probe Design
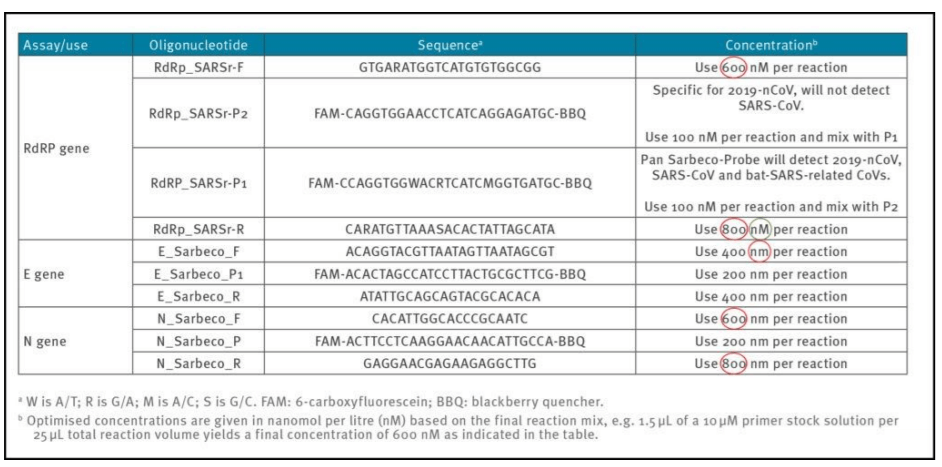
1a) Erroneous primer concentrations
Reliable and accurate PCR−test protocols are normally designed using between 100 nM and 200 nM per primer [7]. In the Corman−Drosten paper, we observe unusually high and varying primer concentrations for several primers (table 1). For the RdRp_SARSr−F and RdRp_SARSr−R primer pairs, 600 nM and 800 nM are described, respectively. Similarly, for the N_Sarbeco_F and N_Sarbeco_R primer set, they advise 600 nM and 800 nM, respectively [1].
It should be clear that these concentrations are far too high to be optimal for specific amplifications of target genes. There exists no specified reason to use these extremely high concentrations of primers in this protocol. Rather, these concentrations lead to increased unspecific binding and PCR product amplification.
Table1: Primers and probes (adapted from Corman−Drosten paper; erroneous primer concentrations are highlighted)
1b) Unspecified (“Wobbly”) primer and probe sequences
To obtain reproducible and comparable results, it is essential to distinctively define the primer pairs. In the Corman−Drosten paper we observed six unspecified positions, indicated by the letters R, W, M and S (Table 2). The letter W means that at this position there can be either an A or a T; R signifies there can be either a G or an A; M indicates that the position may either be an A or a C; the letter S indicates there can be either a G or a C on this position. This high number of variants not only is unusual, but it also is highly confusing for laboratories. These six unspecified positions could easily result in the design of several different alternative primer sequences which do not relate to SARS−CoV−2 (2 distinct RdRp_SARSr_F primers + 8 distinct RdRp_SARS_P1 probes + 4 distinct RdRp_SARSr_R). The design variations will inevitably lead to results that are not even SARS CoV−2 related.
Therefore, the confusing unspecific description in the Corman−Drosten paper is not suitable as a Standard Operational Protocol. These unspecified positions should have been designed unequivocally.
These wobbly sequences have already created a source of concern in the field and resulted in a Letter to the Editor authored by Pillonel et al. [8] regarding blatant errors in the described sequences. These errors are self−evident in the Corman et al. supplement as well.
Table 2: Primers and probes (adapted from Corman−Drosten paper; unspecified (“Wobbly”) nucleotides in the primers are highlighted)
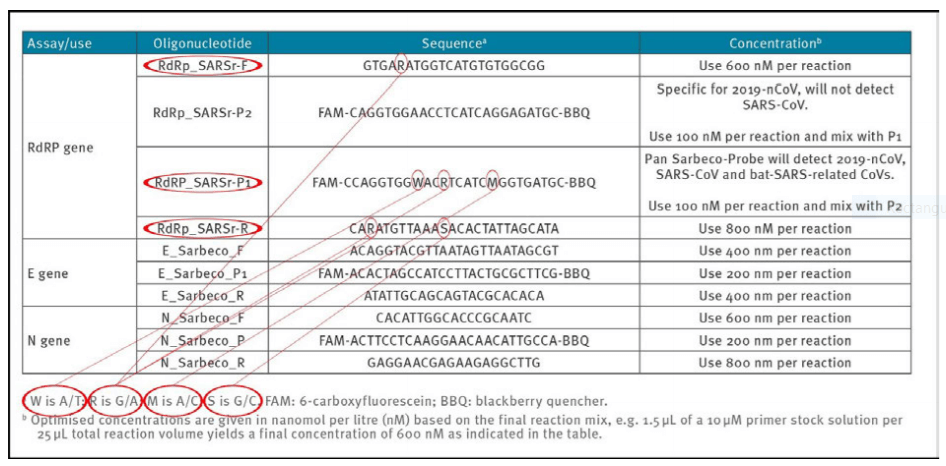
The WHO−protocol (Figure 1), which directly derives from the Corman−Drosten paper, concludes that in order to confirm the presence of SARS−CoV−2, two control genes (the E−and the RdRp−genes) must be identified in the assay. It should be noted, that the RdPd−gene has one uncertain position (“wobbly”) in the forward−primer (R=GƒA), two uncertain positions in the reverse−primer (R=GƒA; S=GƒC) and it has three uncertain positions in the RdRp−probe (W=AƒT; R=GƒA; M=AƒC). So, two different forward primers,
four different reverse primers, and eight distinct probes can be synthesized for the RdPd−gene. Together, there are 64 possible combinations of primers and probes!
The Corman−Drosten paper further identifies a third gene which, according to the WHO protocol, was not further validated and deemed unnecessary:
“Of note, the N gene assay also performed well but was not subjected to intensive further validation because it was slightly less sensitive.”
This was an unfortunate omission as it would be best to use all three gene PCRs as confirmatory assays, and this would have resulted in an almost sufficient virus RNA detection diagnostic tool protocol. Three confirmatory assay−steps would at least minimize−out errors & uncertainties at every fold−step in regards to “Wobbly”−spots. (Nonetheless, the protocol would still fall short of any “good laboratory practice”, when factoring in all the other design−errors).
As it stands, the N gene assay is regrettably neither proposed in the WHO−recommendation (Figure 1) as a mandatory and crucial third confirmatory step, nor is it emphasized in the Corman−Drosten paper as important optional reassurance “for a routine workflow” (Table 2).
Consequently, in nearly all test procedures worldwide, merely 2 primer matches were used instead of all three. This oversight renders the entire test−protocol useless with regards to delivering accurate test−results of real significance in an ongoing pandemic.
Figure 1: The N−Gene confirmatory−assay is neither emphasized as necessary third step in the official WHO Drosten−Corman protocol−recommendation below [8] nor is it required as a crucial step for higher test−accuracy in the Eurosurveillance publication.
1c) Erroneous GC-content (discussed in 2c, together with annealing temperature (Tm))
1d) Detection of viral genes
RT−PCR is not recommended for primary diagnostics of infection. This is why the RT−PCR Test used in clinical routine for detection of COVID−19 is not indicated for COVID−19 diagnosis on a regulatory basis.
“Clinicians need to recognize the enhanced accuracy and speed of the molecular diagnostic techniques for the diagnosis of infections, but also to understand their limitations. Laboratory results should always be interpreted in the context of the clinical presentation of the patient, and appropriate site, quality, and timing of specimen collection are required for reliable test results”. [9]
However, it may be used to help the physician’s differential diagnosis when he or she has to discriminate between different infections of the lung (Flu, Covid−19 and SARS have very similar symptoms). For a confirmative diagnosis of a specific virus, at least 3 specific primer pairs must be applied to detect 3 virus−specific genes. Preferably, these target genes should be located with the greatest distance possible in the viral genome (opposite ends included).
Although the Corman−Drosten paper describes 3 primers, these primers only cover roughly half of the virus’ genome. This is another factor that decreases specificity for detection of intact COVID−19 virus RNA and increases the quote of false positive test results.
Therefore, even if we obtain three positive signals (i.e. the three primer pairs give 3 different amplification products) in a sample, this does not prove the presence of a virus. A better primer design would have terminal primers on both ends of the viral genome. This is because the whole viral genome would be covered and three positive signals can better discriminate between a complete (and thus potentially infectious) virus and fragmented viral genomes (without infectious potency). In order to infer anything of significance about the infectivity of the virus, the Orf1 gene, which encodes the essential replicase enzyme of SARS−CoV viruses, should have been included as a target (Figure 2). The positioning of the targets in the region of the viral genome that is most heavily and variably transcribed is another weakness of the protocol.
Kim et al. demonstrate a highly variable 3’ expression of subgenomic RNA in Sars−CoV−2 [23]. These RNAs are actively monitored as signatures for asymptomatic and non−infectious patients [10]. It is highly questionable to screen a population of asymptomatic people with qPCR primers that have 6 base pairs primer−dimer on the 3 prime end of a primer (Figure 3).
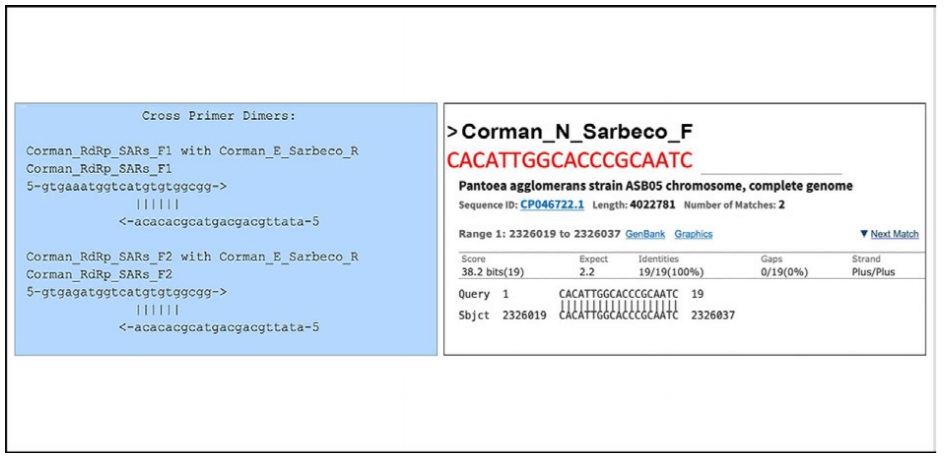
Apparently the WHO recommends these primers. We tested all the wobble derivatives from the Corman−Drosten paper with Thermofisher’s primer dimer web tool [11]. The RdRp forward primer has 6bp 3prime homology with Sarbeco E Reverse. At high primer concentrations this is enough to create inaccuracies.
Of note: There is a perfect match of one of the N primers to a clinical pathogen (Pantoea), found in immuno−compromised patients. The reverse primer hits Pantoea as well but not in the same region (Figure 3).
These are severe design errors, since the test cannot discriminate between the whole virus and viral fragments. The test cannot be used as a diagnostic for SARS−viruses.
Figure 2: Relative positions of amplicon targets on the SARS coronavirus and the 2019 novel coronavirus genome. ORF: open reading frame; RdRp: RNA−dependent RNA polymerase. Numbers below amplicon are genome positions according to SARS−CoV, NC_004718 [1];
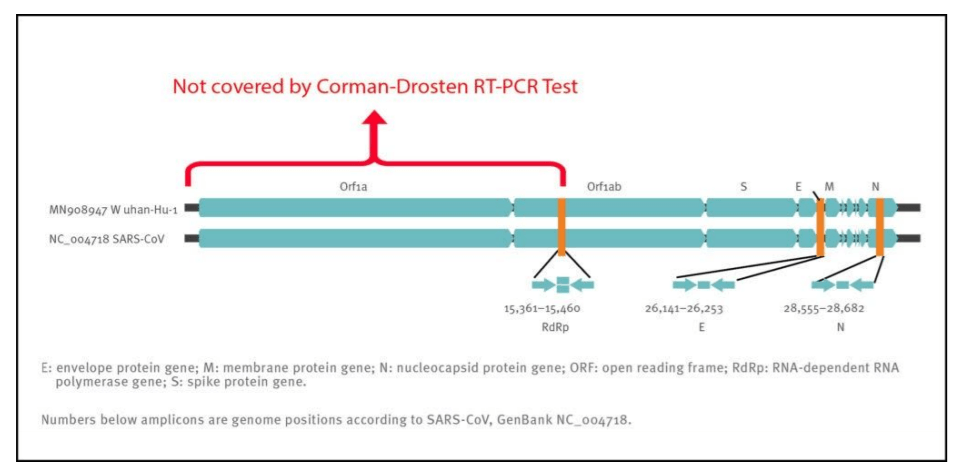
Figure 3: A test with Thermofischer’s primer dimer web tool reveals that the RdRp forward primer has a 6bp 3`prime homology with Sarbeco E Reverse (left box). Another test reveals that there is a perfect match for one of the N−primers to a clinical pathogen (Pantoea) found in immuno−compromised patients (right box).
2. Reaction temperature
2a) DNA melting temperature (>92˚).
Adequately addressed in the Corman−Drosten paper.
2b) DNA amplification temperature.
Adequately addressed in the Corman−Drosten paper.
2c) Erroneous GC−contents and Tm
The annealing−temperature determines at which temperature the primer attachesƒdetaches from the target sequence. For an efficient and specific amplification, GC content of primers should meet a minimum of 40% and a maximum of 60% amplification. As indicated in table 3, three of the primers described in the Corman−Drosten paper are not within the normal range for GC−content. Two primers (RdRp_SARSr_F and RdRp_SARSr_R) have unusual and very low GC−values of 28%−31% for all possible variants of wobble bases, whereas primer E_Sarbeco_F has a GC−value of 34.6% (Table 3 and second panel of Table 3).
It should be noted that the GC−content largely determines the binding to its specific target due to its three hydrogen bonds in base pairing. Thus, the lower the GC−content of the primer, the lower its binding−capability to its specific target gene sequence (i.e. the gene to be detected). This means for a target−sequence to be recognized we have to choose a temperature which is as close as possible to the actual annealing−temperature (best practise−value) for the primer not to detach again, while at the same time specifically selecting the target sequence.
If the Tm−value is very low, as observed for all wobbly−variants of the RdRp reverse primers, the primers can bind non−specifically to several targets, decreasing specificity and increasing potential false positive results.
The annealing temperature (Tm) is a crucial factor for the determination of the specificityƒaccuracy of the qPCR procedure and essential for evaluating the accuracy of qPCR−protocols. Best−practice recommendation: Both primers (forward and reverse) should have an almost similar value, preferably the identical value.
We used the freely available primer design software Primer−BLAST [12, 25] to evaluable the best−practise values for all primers used in the Corman−Drosten paper (Table 3). We attempted to find a Tm−value of 60˚ C, while similarly seeking the highest possible GC%−value for all primers. A maximal Tm difference of 2˚ C within primer pairs was considered acceptable. Testing the primer pairs specified in the Corman−Drosten paper, we observed a difference of 10˚ C with respect to the annealing temperature Tm for primer pair1 (RdRp_SARSr_F and RdRp_SARSr_R). This is a very serious error and makes the protocol useless as a specific diagnostic tool.
Additional testing demonstrated that only the primer pair designed to amplify the N−gene (N_Sarbeco_F and N_Sarbeco_R) reached the adequate standard to operate in a diagnostic test, since it has a sufficient GC−content and the Tm difference between the primers (N_Sarbeco_F and N_Sarbeco_R) is 1.85˚ C (below the crucial maximum of 2˚ C difference). Importantly, this is the gene which was neither tested in the virus samples (Table 2) nor emphasized as a confirmatory test. In addition to highly variable melting temperatures and degenerate sequences in these primers, there is another factor impacting specificity of the procedure: the dNTPs (0.4uM) are 2x higher than recommended for a highly specific amplification. There is additional magnesium sulphate added to the reaction as well. This procedure combined with a low annealing temperature can create non−specific amplifications. When additional magnesium is required for qPCR, specificity of the assay should be further scrutinized.
The design errors described here are so severe that it is highly unlikely that specific amplification of SARS−CoV−2 genetic material will occur using the protocol of the Corman−Drosten paper.
Table 3: GC−content of the primers and probes (adapted from Corman−Drosten paper; aberrations from optimized GC−contents are highlighted. Second Panel shows a table−listing of all Primer−BLAST best practices values for all primers and probes used in the Corman−Drosten paper by Prof. Dr. Ulrike Kämmerer & her team.
3. The number of amplification cycles
It should be noted that there is no mention anywhere in the Corman−Drosten paper of a test being positive or negative, or indeed what defines a positive or negative result. These types of virological diagnostic tests must be based on a SOP, including a validated and fixed number of PCR cycles (Ct value) after which a sample is deemed positive or negative. The maximum reasonably reliable Ct value is 30 cycles. Above a Ct of 35 cycles, rapidly increasing numbers of false positives must be expected .
PCR data evaluated as positive after a Ct value of 35 cycles are completely unreliable.
Citing Jaafar et al. 2020 [3]:
“At Ct = 35, the value we used to report a positive result for PCR, <3% of cultures are positive.”
In other words, there was no successful virus isolation of SARS−CoV−2 at those high Ct values. Further, scientific studies show that only non−infectious (dead) viruses are detected with Ct values of 35 [22].
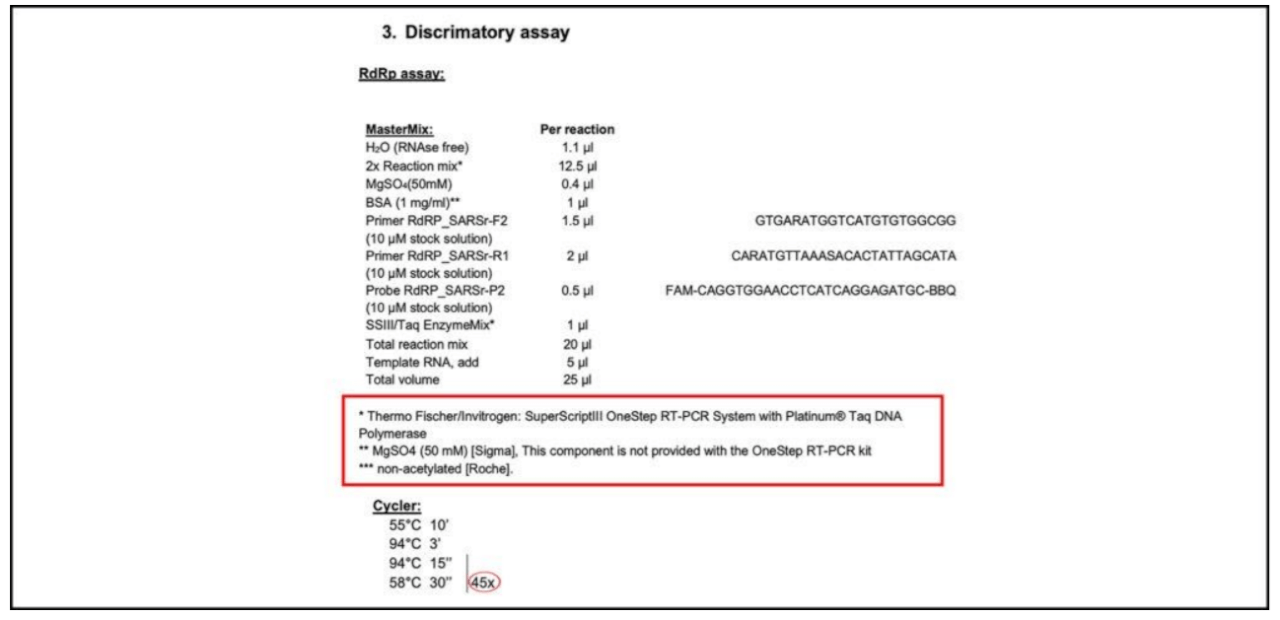
Between 30 and 35 there is a grey area, where a positive test cannot be established with certainty. This area should be excluded. Of course, one could perform 45 PCR cycles, as recommended in the Corman−Drosten WHO−protocol (Figure 4), but then you also have to define a reasonable Ct−value (which should not exceed 30). But an analytical result with a Ct value of 45 is scientifically and diagnostically absolutely meaningless (a reasonable Ct−value should not exceed 30). All this should be communicated very clearly. It is a significant mistake that the Corman−Drosten paper does not mention the maximum Ct value at which a sample can be unambiguously considered as a positive or a negative test−result. This important cycle threshold limit is also not specified in any follow−up submissions to date.
Figure 4: RT−PCR Kit recommendation in the official Corman−Drosten WHO−protocol [8]. Only a “Cycler”−value (cycles) is to be found without corresponding and scientifically reasonable Ct (Cutoff−value). This or any other cycles−value is nowhere to be found in the actual Corman−Drosten paper.
4. Biomolecular validations
To determine whether the amplified products are indeed SARS−CoV−2 genes, biomolecular validation of amplified PCR products is essential. For a diagnostic test, this validation is an absolute must.
Validation of PCR products should be performed by either running the PCR product in a 1% agarose−EtBr gel together with a size indicator (DNA ruler or DNA ladder) so that the size of the product can be estimated. The size must correspond to the calculated size of the amplification product. But it is even better to sequence the amplification product. The latter will give 100% certainty about the identity of the amplification product. Without molecular validation one can not be sure about the identity of the amplified PCR products. Considering the severe design errors described earlier, the amplified PCR products can be anything.
Also not mentioned in the Corman−Drosten paper is the case of small fragments of qPCR (around 100bp): It could be either 1,5% agarose gel or even an acrylamide gel.
The fact that these PCR products have not been validated at molecular level is another striking error of the protocol, making any test based upon it useless as a specific diagnostic tool to identify the SARS−CoV−2 virus.
5. Positive and negative controls to confirm/refute specific virus detection.
The unconfirmed assumption described in the Corman−Drosten paper is that SARS−CoV−2 is the only virus from the SARS−like beta−coronavirus group that currently causes infections in humans. The sequences on which their PCR method is based are in silico sequences, supplied by a laboratory in China [23], because at the time of development of the PCR test no control material of infectious (“live”) or inactivated SARS−CoV−2 was available to the authors. The PCR test was therefore designed using the sequence of the known SARS−CoV as a control material for the Sarbeco component (Dr. Meijer, co−author Corman−Drosten paper in an email exchange with Dr. Peter Borger) [2].
All individuals testing positive with the RT−PCR test, as described in the Corman−Drosten paper, are assumed to be positive for SARS−CoV−2 infections. There are three severe flaws
in their assumption. First, a positive test for the RNA molecules described in the Corman−Drosten paper cannot be equated to “infection with a virus”. A positive RT−PCR test merely indicates the presence of viral RNA molecules. As demonstrated under point 1d (above), the Corman−Drosten test was not designed to detect the full−length virus, but only a fragment of the virus. We already concluded that this classifies the test as unsuitable as a diagnostic test for SARS−virus infections.
Secondly and of major relevance, the functionality of the published RT−PCR Test was not demonstrated with the use of a positive control (isolated SARS−CoV−2 RNA) which is an essential scientific gold standard.
Third, the Corman−Drosten paper states:
“To show that the assays can detect other bat−associated SARS−related viruses, we used the E gene assay to test six bat−derived faecal samples available from Drexler et al. […] und Muth et al. […]. These virus−positive samples stemmed from European rhinolophid bats. Detection of these phylogenetic outliers within the SARS−related CoV clade suggests that all Asian viruses are likely to be detected. This would, theoretically, ensure broad sensitivity even in case of multiple independent acquisitions of variant viruses from an animal reservoir.”
This statement demonstrates that the E gene used in RT−PCR test, as described in the Corman−Drosten paper, is not specific to SARS−CoV−2.
The E gene primers also detect a broad spectrum of other SARS viruses.
The genome of the coronavirus is the largest of all RNA viruses that infect humans and they all have a very similar molecular structure. Still, SARS−CoV1 and SARS−CoV−2 have two highly specific genetic fingerprints, which set them apart from the other coronaviruses. First, a unique fingerprint−sequence (KTFPPTEPKKDKKKK) is present in the N−protein of SARS−CoV and SARS−CoV−2 [13,14,15]. Second, both SARS−CoV1 and SARS−CoV2 do not contain the HE protein, whereas all other coronaviruses possess this gene [13, 14]. So, in order to specifically detect a SARS−CoV1 and SARS−CoV−2 PCR product the above region in the N gene should have been chosen as the amplification target. A reliable diagnostic test should focus on this specific region in the N gene as a confirmatory test. The PCR for this N gene was not further validated nor recommended as a test gene by the Drosten−Corman paper, because of being “not so sensitive” with the SARS−CoV original probe [1].
Furthermore, the absence of the HE gene in both SARS−CoV1 and SARS−CoV−2 makes this gene the ideal negative control to exclude other coronaviruses. The Corman−Drosten paper does not contain this negative control, nor does it contain any other negative controls. The
PCR test in the Corman−Drosten paper therefore contains neither a unique positive control nor a negative control to exclude the presence of other coronaviruses. This is another major design flaw which classifies the test as unsuitable for diagnosis.
6. Standard Operational Procedure (SOP) is not available
There should be a Standard Operational Procedure (SOP) available, which unequivocally specifies the above parameters, so that all laboratories are able to set up the identical same test conditions. To have a validated universal SOP is essential, because it facilitates data comparison within and between countries. It is very important to specify all primer parameters unequivocally. We note that this has not been done. Further, the Ct value to indicate when a sample should be considered positive or negative is not specified. It is also not specified when a sample is considered infected with SARS−CoV viruses. As shown above, the test cannot discern between virus and virus fragments, so the Ct value indicating positivity is crucially important. This Ct value should have been specified in the Standard Operational Procedure (SOP) and put on−line so that all laboratories carrying out this test have exactly the same boundary conditions. It points to flawed science that such an SOP does not exist. The laboratories are thus free to conduct the test as they consider appropriate, resulting in an enormous amount of variation. Laboratories all over Europe are left with a multitude of questions; which primers to order? which nucleotides to fill in the undefined places? which Tm value to choose? How many PCR cycles to run? At what Ct value is the sample positive? And when is it negative? And how many genes to test? Should all genes be tested, or just the E and RpRd gene as shown in Table 2 of the Corman−Drosten paper? Should the N gene be tested as well? And what is their negative control? What is their positive control?
The protocol as described is unfortunately very vague and erroneous in its design that one can go in dozens of different directions. There does not appear to be any standardization nor an SOP, so it is not clear how this test can be implemented.
7. Consequences of the errors described under 1-5: false positive results.
The RT−PCR test described in the Corman−Drosten paper contains so many molecular biological design errors (see 1−5) that it is not possible to obtain unambiguous results. It is inevitable that this test will generate a tremendous number of so−called “false positives”. The definition of false positives is a negative sample, which initially scores positive, but which is negative after retesting with the same test. False positives are erroneous positive test−results, i.e. negative samples that test positive. And this is indeed what is found in the Corman−Drosten paper. On page 6 of the manuscript PDF the authors demonstrate, that even under well−controlled laboratory conditions, a considerable percentage of false positives is generated with this test:
“In four individual test reactions, weak initial reactivity was seen however they were negative upon retesting with the same assay. These signals were not associated with any particular virus, and for each virus with which initial positive reactivity occurred, there were other samples that contained the same virus at a higher concentration but did not test positive.
Given the results from the extensive technical qualification described above, it was concluded that this initial reactivity was not due to chemical instability of real−time PCR probes and most probably to handling issues caused by the rapid introduction of new diagnostic tests and controls during this evaluation study.” [1]
The first sentence of this excerpt is clear evidence that the PCR test described in the Corman−Drosten paper generates false positives. Even under the well−controlled conditions of the state−of−the−art Charité−laboratory, 4 out of 310 primary−tests are false positives per definition. Four negative samples initially tested positive, then were negative upon retesting. This is the classical example of a false positive. In this case the authors do not identify them as false positives, which is intellectually dishonest.
Another telltale observation in the excerpt above is that the authors explain the false positives away as “handling issues caused by the rapid introduction of new diagnostic tests”. Imagine the laboratories that have to introduce the test without all the necessary information normally described in an SOP.
8. The Corman-Drosten paper was not peer-reviewed
Before formal publication in a scholarly journal, scientific and medical articles are traditionally certified by “peer review.” In this process, the journal’s editors take advice from various experts (“referees”) who have assessed the paper and may identify weaknesses in its assumptions, methods, and conclusions. Typically a journal will only publish an article once the editors are satisfied that the authors have addressed referees’ concerns and that the data presented supports the conclusions drawn in the paper.” This process is as well described for Eurosurveillance [16].
The Corman−Drosten paper was submitted to Eurosurveillance on January 21st 2020 and accepted for publication on January 22nd 2020. On January 23rd 2020 the paper was online. On January 13th 2020 version 1−0 of the protocol was published at the official WHO website [17], updated on January 17th 2020 as document version 2−1 [18], even before the Corman−Drosten paper was published on January 23rd at Eurosurveillance.
Normally, peer review is a time−consuming process since at least two experts from the field have to critically read and comment on the submitted paper. In our opinion, this paper was not peer−reviewed. Twenty−four hours are simply not enough to carry out a thorough peer review. Our conclusion is supported by the fact that a tremendous number of very serious design flaws were found by us, which make the PCR test completely unsuitable as a diagnostic tool to identify the SARS−CoV−2 virus. Any molecular biologist familiar with RT−PCR
design would have easily observed the grave errors present in the Corman−Drosten paper before the actual review process. We asked Eurosurveillance on October 26th 2020 to send us a copy of the peer review report. To date, we have not received this report and in a letter dated November 18th 2020, the ECDC as host for Eurosurveillance declined to provide access without providing substantial scientific reasons for their decision. On the contrary, they write that “disclosure would undermine the purpose of scientific investigations.” [24].
9. Authors as the editors
A final point is one of major concern. It turns out that two authors of the Corman−Drosten paper, Christian Drosten and Chantal Reusken, are also members of the editorial board of this journal [19]. Hence there is a severe conflict of interest which strengthens suspicions that the paper was not peer−reviewed. It has the appearance that the rapid publication was possible simply because the authors were also part of the editorial board at Eurosurveillance. This practice is categorized as compromising scientific integrity.
SUMMARY CATALOGUE OF ERRORS FOUND IN THE PAPER
EXPANDThe Corman−Drosten paper contains the following specific errors:
- There exists no specified reason to use these extremely high concentrations of primers in this protocol. The described concentrations lead to increased nonspecific bindings and PCR product amplifications, making the test unsuitable as a specific diagnostic tool to identify the SARS−CoV−2
- Six unspecified wobbly positions will introduce an enormous variability in the real world laboratory implementations of this test; the confusing nonspecific description in the Corman−Drosten paper is not suitable as a Standard Operational Protocol making the test unsuitable as a specific diagnostic tool to identify the SARS−CoV−2
- The test cannot discriminate between the whole virus and viral fragments. Therefore, the test cannot be used as a diagnostic for intact (infectious) viruses, making the test unsuitable as a specific diagnostic tool to identify the SARS−CoV−2 virus and make inferences about the presence of an
- A difference of 10˚ C with respect to the annealing temperature Tm for primer pair1 (RdRp_SARSr_F and RdRp_SARSr_R) also makes the test unsuitable as a specific diagnostic tool to identify the SARS−CoV−2
- A severe error is the omission of a Ct value at which a sample is considered positive and negative. This Ct value is also not found in follow−up submissions making the test unsuitable as a specific diagnostic tool to identify the SARS−CoV−2
- The PCR products have not been validated at the molecular level. This fact makes the protocol useless as a specific diagnostic tool to identify the SARS−CoV−2
- The PCR test contains neither a unique positive control to evaluate its specificity for SARS−CoV−2 nor a negative control to exclude the presence of other coronaviruses, making the test unsuitable as a specific diagnostic tool to identify the SARS−CoV−2
- The test design in the Corman−Drosten paper is so vague and flawed that one can go in dozens of different directions; nothing is standardized and there is no SOP. This highly questions the scientific validity of the test and makes it unsuitable as a specific diagnostic tool to identify the SARS−CoV−2
- Most likely, the Corman−Drosten paper was not peer−reviewed making the test unsuitable as a specific diagnostic tool to identify the SARS−CoV−2
- We find severe conflicts of interest for at least four authors, in addition to the fact that two of the authors of the Corman−Drosten paper (Christian Drosten and Chantal Reusken) are members of the editorial board of Eurosurveillance. A conflict of interest was added on July 29 2020 (Olfert Landt is CEO of TIB−Molbiol; Marco Kaiser is senior researcher at GenExpress and serves as scientific advisor for TIB−Molbiol), that was not declared in the original version (and still is missing in the PubMed version); TIB−Molbiol is the company which was “the first” to produce PCR kits (Light Mix) based on the protocol published in the Corman−Drosten manuscript, and according to their own words, they distributed these PCR−test kits before the publication was even submitted [20]; further, Victor Corman & Christian Drosten failed to mention their second affiliation: the commercial test laboratory “Labor Berlin”. Both are responsible for the virus diagnostics there [21] and the company operates in the realm of real time PCR−testing.
In light of our re-examination of the test protocol to identify SARS-CoV-2 described in the Corman-Drosten paper we have identified concerning errors and inherent fallacies which render the SARS-CoV-2 PCR test useless.
CONCLUSION
The decision as to which test protocols are published and made widely available lies squarely in the hands of Eurosurveillance. A decision to recognise the errors apparent in the Corman−Drosten paper has the benefit to greatly minimise human cost and suffering going forward.
Is it not in the best interest of Eurosurveillance to retract this paper? Our conclusion is clear. In the face of all the tremendous PCR−protocol design flaws and errors described here, we conclude: There is not much of a choice left in the framework of scientific integrity and responsibility.
REFERENCES
EXPAND- Corman Victor M, Landt Olfert, Kaiser Marco, Molenkamp Richard, Meijer Adam, Chu Daniel KW, Bleicker Tobias, Brünink Sebastian, Schneider Julia, Schmidt Marie Luisa, Mulders Daphne GJC, Haagmans Bart L, van der Veer Bas, van den Brink Sharon, Wijsman Lisa, Goderski Gabriel, Romette Jean−Louis, Ellis Joanna, Zambon Maria, Peiris Malik, Goossens Herman, Reusken Chantal, Koopmans Marion PG, Drosten Christian. Detection of 2019 novel coronavirus (2019−nCoV) by real−time RT−PCR. Euro Surveill. 2020;25(3):pii=2000045. https:ƒƒdoi.orgƒ10.2807ƒ1560−7917.ES.2020.25.3.2000045
- Email communication between Dr. Peter Borger & Dr. Adam Meijer: Supplementary Material
- Jafaar et , Correlation Between 3790 Quantitative Polymerase Chain Reaction−Positives Samples and Positive Cell Cultures, Including 1941 Severe Acute Respiratory Syndrome Coronavirus 2 Isolates. https:ƒƒacademic.oup.comƒcidƒadvance−articleƒdoiƒ10.1093ƒcidƒciaa1491ƒ5912603
- BBC, January 21st 2020: https:ƒƒbbc.comƒnewsƒworld−asia−china−51185836;
Archive: https:ƒƒarchive.isƒ0qRmZ
- Google Analytics − COVID19−deaths worldwide: https:ƒƒbit.lyƒ3fndemJ
Archive: https:ƒƒarchive.isƒPpqEE
- Laboratory testing for COVID−19 Emergency Response Technical Centre, NIVD under
China CDC March 15th, 2020: http:ƒƒwww.chinacdc.cnƒenƒCOVID19ƒ202003ƒP020200323390321297894.pdf
- Real−Time PCR Handbook Life Technologies: https:ƒƒthermofisher.comƒcontentƒdamƒLifeTechƒglobalƒFormsƒPDFƒreal−time−pcr−
handbook.pdf
Nolan T, Huggett J, Sanchez E.Good practice guide for the application of quantitative PCR (qPCR) First Edition 2013
- Trestan Pillonel et al, Letter to the editor: SARS−CoV−2 detection by real−time RT−PCR: https:ƒƒncbi.nlm.nih.govƒpmcƒarticlesƒPMC7268274ƒ
- Kurkela, Satu, and David WG “Molecular−diagnostic techniques.” Medicine 38.10 (2009): 535−540.
- Wolfel et , Virological assessment of hospitalized patients with COVID−2019 https:ƒƒwww.nature.comƒarticlesƒs41586−020−2196−x
- Thermofischer Primer Dimer Web Tool: https:ƒƒthermofisher.comƒusƒenƒhomeƒbrandsƒthermo−scientificƒmolecular−biologyƒmolecular−biology
−learning−centerƒmolecular−biology−resource−libraryƒthermo−scientific−web−toolsƒmultiple−primer−analyzer.h tml
Suplementary material by Kevin Mckernan, Corman−Drosten Primer Dimer Results with Thermofischer Primer Dimer Web Tool
- Primer−BLAST, NCBI − National Center for Biotechnology Information: https:ƒƒncbi.nlm.nih.govƒtoolsƒprimer−blastƒ
- Marra MA, Steven JMJ, Caroline RA, Robert AH, Angela BW et (2003) Science. The Genome sequence of the SARS−associated coronavirus. Science 300(5624): 1399−1404.
- Severe acute respiratory syndrome coronavirus 2 isolate Wuhan−Hu−1, complete genome: https:ƒƒncbi.nlm.nih.govƒnuccoreƒMN908947
- Borger A SARS−like Coronavirus was expected but nothing was done to be prepared. Am J Biomed Sci Res 2020. https:ƒƒbiomedgrid.comƒpdfƒAJBSR.MS.ID.001312.pdf
https:ƒƒwww.researchgate.netƒpublicationƒ341120750_A_SARS−like_Coronavirus_was_Expected_but_nothi ng_was_done_to_be_Prepared;
Archive: https:ƒƒarchive.isƒi76Hu
- Eurosurveillance paper evaluation ƒ review process: https:ƒƒeurosurveillance.orgƒevaluation
- Official recommendation of the Corman−Drosten protocol & manuscript by the WHO,published on January 13th 2020 as version 1.0 of the document:
https:ƒƒwww.who.intƒdocsƒdefault−sourceƒcoronaviruseƒwuhan−virus−assay−v1991527e5122341d99287a1b1 7c111902.pdf; archive: https:ƒƒbit.lyƒ3m3jXVH
- Official WHO−recommendation for the Corman ƒ Drosten RT−qPCR−protocol, which directly derives from the Eurosurveillance−publication, document−version 2−1, published on 17th January 2020: https:ƒƒwww.who.intƒdocsƒdefault−sourceƒcoronaviruseƒprotocol−v2−1.pdf?sfvrsn=a9ef618c_2
- Eurosurveillance Editorial Board, 2020: https:ƒƒeurosurveillance.orgƒuploadƒsite−assetsƒimgsƒ2020−09−Editorial%20Board%20PDF.pdf;
Archive: https:ƒƒbit.lyƒ2TqXBjX
- Instructions For Use LightMix SarbecoV E−gene plus EAV Control, TIB−Molbiol & Roche Molecular Solutions, January 11th 2020: https:ƒƒwww.roche−as.esƒlm_pdfƒMDx_40−0776_96_Sarbeco−E−gene_V200204_09164154001 (1).pdf
Archive, timestamp − January 11th 2020: https:ƒƒarchive.isƒVulo5;
Archive: https:ƒƒbit.lyƒ3fm9bXH
- Christian Drosten & Victor Corman, responsible for viral diagnostics at Labor Berlin: https:ƒƒlaborberlin.comƒfachbereicheƒvirologieƒ
Archive: https:ƒƒarchive.isƒCDEUG
- Tom Jefferson, Elizabeth Spencer, Jon Brassey, Carl Heneghan Viral cultures for COVID−19 infectivity assessment. Systematic review. Systematic review doi: https:ƒƒdoi.orgƒ10.1101ƒ2020.08.04.20167932 https:ƒƒmedrxiv.orgƒcontentƒ10.1101ƒ2020.08.04.20167932v4
- Kim et al.,The Architecture of SARS−CoV−2 Transcriptome: https:ƒƒsciencedirect.comƒscienceƒarticleƒpiiƒS0092867420304062
- ECDC reply to Dr. Peter Borger, 18th November 2020: Supplementary Material
- Dr. Ulrike Kämmerer & team, survey & Primer−BLAST table: Supplementary Material
Additional literature:
Description RT−PCR RKI Germany, on page 10 of this link: https:ƒƒwww.rki.deƒDEƒContentƒGesundheitsmonitoringƒGesundheitsberichterstattungƒGBEDownloadsJƒJo HM_S5_2020_Studienprotokoll_CORONA_MONITORING_lokal.pdf? blob=publicationFile
Author’s Affiliations:
EXPAND- Pieter Borger (MSc, PhD), Molecular Genetics, W+W Research Associate, Lörrach, Germany
- Rajesh Kumar Malhotra (Artist Alias: Bobby Rajesh Malhotra), Former 3D Artist / Scientific Visualizations at CeMM – Center for Molecular Medicine of the Austrian Academy of Sciences (2019-2020), University for Applied Arts – Department for Digital Arts Vienna, Austria
- Michael Yeadon BSs(Hons) Biochem Tox U Surrey, PhD Pharmacology U Surrey. Managing Director, Yeadon Consulting Ltd, former Pfizer Chief Scientist, United Kingdom
- Clare Craig MA, (Cantab) BM, BCh (Oxon), FRCPath, United Kingdom
- Kevin McKernan, BS Emory University, Chief Scientific Officer, founder Medical Genomics, engineered the sequencing pipeline at WIBR/MIT for the Human Genome Project, Invented and developed the SOLiD sequencer, awarded patents related to PCR, DNA Isolation and Sequencing, USA
- Dr. Klaus Steger, Department of Urology, Pediatric Urology and Andrology, Molecular Andrology, Biomedical Research Center of the Justus Liebig University, Giessen, Germany
- Paul McSheehy (BSc, PhD), Biochemist & Industry Pharmacologist, Loerrach, Germany
- Lidiya Angelova, MSc in Biology, PhD in Microbiology, Former researcher at the National Institute of Allergy and Infectious Diseases (NIAID), Maryland, USA
- Fabio Franchi, Former Dirigente Medico (M.D) in an Infectious Disease Ward, specialized in “Infectious Diseases” and “Hygiene and Preventive Medicine”, Società Scientifica per il Principio di Precauzione (SSPP), Italy
- med. Thomas Binder, Internist and Cardiologist (FMH), Switzerland
- Dr. med. Henrik Ullrich, specialist Diagnostic Radiology, Chief Medical Doctor at the Center for Radiology of Collm Oschatz-Hospital, Germany
- Dr. Makoto Ohashi, Professor emeritus, PhD in Microbiology and Immunology, Tokushima University, Japan
- Stefano Scoglio, B.Sc. Ph.D., Microbiologist, Nutritionist, Italy
- Marjolein Doesburg-van Kleffens (MSc, PhD), specialist in Laboratory Medicine (clinical chemistry), Maasziekenhuis Pantein, Beugen, The Netherlands
- Dorothea Gilbert (MSc, PhD), PhD Environmental Chemistry and Toxicology. DGI Consulting Services, Oslo, Norway
- Rainer J. Klement, PhD. Department of Radiation Oncology, Leopoldina Hospital Schweinfurt, German
- Ruth Schruefer, PhD, human genetics/ immunology, Munich, Germany,
- Berber W. Pieksma, General Practitioner, The Netherlands
- med. Jan Bonte (GJ), Consultant Neurologist, The Netherlands
- Bruno H. Dalle Carbonare (Molecular biologist), IP specialist, BDC Basel, Switzerland
- Kevin P. Corbett, MSc Nursing (Kings College London) PhD (London South Bank) Social Sciences (Science & Technology Studies) London, England, United Kingdom
- Dr. Ulrike Kämmerer, specialist in Virology / Immunology / Human Biology / Cell Biology, University Hospital Würzburg, Germany
Author’s Contributions:
EXPANDPB: Planned and conducted the analyses and research, conceptualising the manuscript. BRM: Planned and conducted the research, conceptualising the figures and manuscript. MY: Conducted the analyses and research.
KMcK: Conducted the analyses and research, conceptualized the manuscript. KS: Conducted the analyses and research.
PMcS: Proofreading the analyses and research.
LA: Proofreading the analyses and research. FF: Proofreading the analyses and research. TB: Proofreading the analyses and research. HU: Proofreading the analyses and research. MO: Proofreading the analyses and research. SS: Proofreading the analyses and research.
MDvK: Proofreading the analyses and research.
DG: Proofreading the analyses and research. RJK: Proofreading the analyses and research.
RS: Proofreading the analyses and research, and the manuscript. BWK: Proofreading the analyses and research.
RvV: Proofreading the analyses and research. JB: Proofreading the analyses and research. KC: Proofreading the analyses and research.
UK: Planned and conducted the analyses and research, conceptualising the manuscript.
Additional Proof-Readers:
EXPANDSaji N Hameed, Environmental Informatics, University of Aizu, Tsuruga, Ikki-machi, Aizuwakamatsu-shi, Fukushima, Japan Howard R. Steen, MA Chem. Eng. Cantab, Former Research Manager, Germany
